Figures & data
Figure 1. Routine blood test and histopathological diagnosis of ApcMin/+ mice ingested different concentration of Neu5Gc. (A) Experimental process of ingested different concentrations of Neu5Gc in ApcMin/+ mice; (B) Histopathological diagnosis of colorectal in ApcMin/+ mice; (C) Representative H&E staining histological sections in the colorectal in different groups: left: Control group; right: Treatment group; top panel: bar = 250 µm; bottom panel: bar = 100 µm; (D) Routine blood analysis of ApcMin/+ mice, red blood cells (RBC), platelets (PLT), Lymphocyte count (Lymph), white blood cells (WBC).
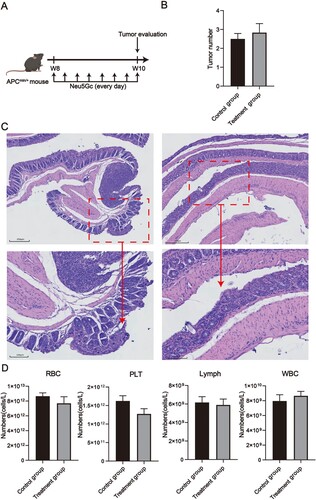
Figure 2. RNA-seq analysis of the colorectal of ApcMin/+ mice ingested different concentration of Neu5Gc. (A) t-SNE plots of relative gene expression in the colorectal; (B) Volcano plot for DEGs; (C) Top 10 GO terms of the up-DEGs; (D) Top 10 enriched pathways of the up-DEGs; (E) Top 10GO terms; (F) Top 10 enriched pathways.
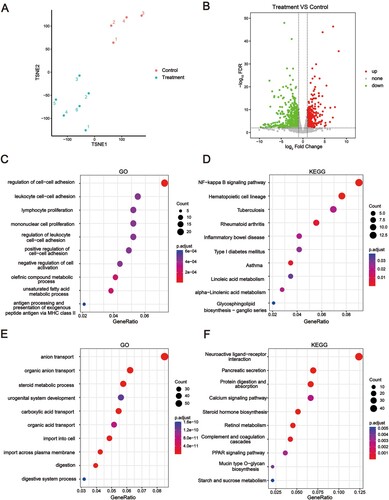
Figure 3. PPI analysis of DEGs. DEGs between Treatment and Control groups were investigated using String. The interaction between each protein pair is represented by a line, and the size of the circle is proportional to the degree of interaction. Proteins that are closer to the concentric circles have higher interactions with other proteins.
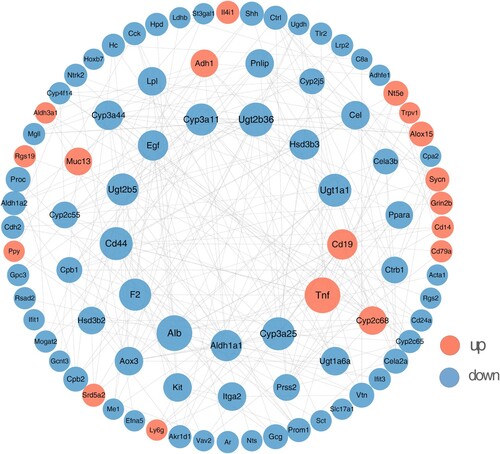
Figure 4. RNA-seq of the liver of ApcMin/+ mice ingested different concentrations of Neu5Gc. (A) Volcano plot of DEGs in livers; (B) Top 10 enriched GO pathways of the up-DEGs involved the liver of ApcMin/+ mice; (C) Top 10 enriched GO pathways of the down-DEGs involved the liver of ApcMin/+ mice.
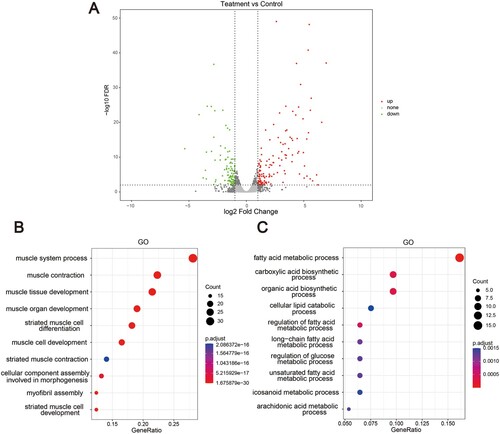
Figure 5. Comparison of the TPM value and RT-qPCR of the Cmah gene in the colorectal and liver in the Treatment group and the Control group. (A) Histogram comparing the TPM value of the Cmah gene in the colorectal Treatment group and Control group; (B) The relative expression of the Cmah gene in the RT-qPCR results was compared between the colorectal Treatment group and the Control group; (C) Histogram comparing the TPM value of Cmah gene in the Treatment group and the Control group in the liver; (D) Comparison of the relative expression of Cmah gene in the RT-qPCR results of liver Treatment group and Control group. * indicates p < 0.05, ** indicates p < 0.01.
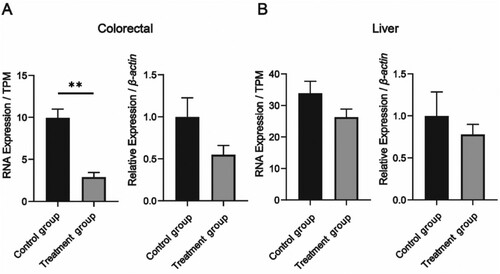
Table 1. The Oligonucleotide primers used in RT-qPCR analysis.
Supplemental Material
Download MS Excel (11.9 MB)Data availability statement
Data is contained within the article.
