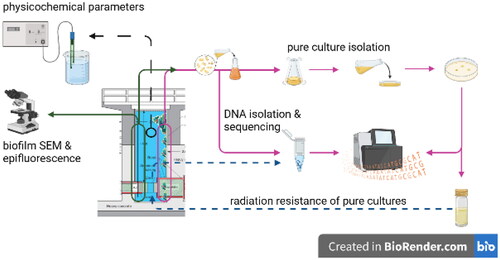
Figures & data
Figure 1. (A) Side view of the JSI TRIGA Mark II research reactor. The ladder with sampling slides was positioned close to wall on the left-hand side of this schematic, indicated by a full black vertical line. See for photograph of the glass slides. (B) Schematic figure of the JSI TRIGA reactor core. Channel F26 is located in the outer ring of JSI TRIGA core (black arrow).
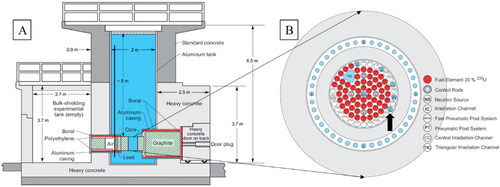
Figure 2. JSI TRIGA research reactor pool with ladder and attached glass slides. To the left of the ladder is the reactor core. Biofilm growing on the wall itself can be observed. Red arrows indicate position of the biofilm sampling glass slides. Photograph taken during glass-slide deployments on the 30 December 2014. See caption for placement information.
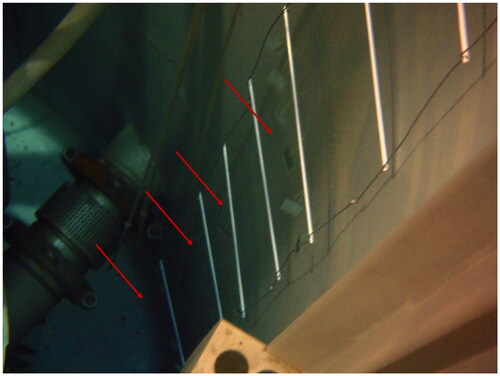
Table 1. Measured doses by depth at constant reactor power (1 kW).
Table 2. Chemical conditions on the 16 January 2014, at the beginning and at the end of first the biofilm sampling.
Table 3. Stable isotope analysis of reactor water and biofilm.
Table 4. Results of ICP-MS screening of reactor biofilm.
Table 5. Visual characteristics of pure cultures chosen for 16S rDNA Sanger sequencing.
Table 6. Overview of samples for metagenomic 16S rDNA sequencing.
Figure 3. Epifluorescence micrographs of native biofilm under excitation with UV illumination corresponding to autofluorescence of chlorophyll a. A 3D composite image (not shown) showed the chlorophyll was distributed throughout the biofilm, meaning there was no spatial segregation of the phototrophs.
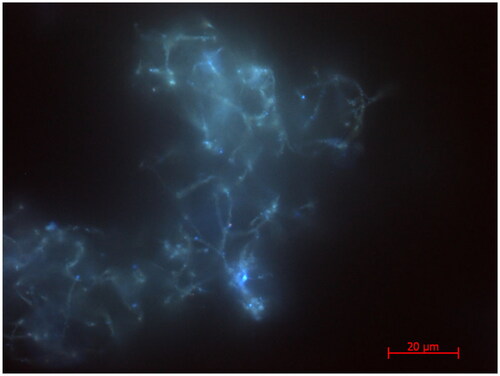
Figure 4. Colony of B. flexus, after irradiation with γ + n0. Agglomeration in sphere-like structure was not anticipated. No further growth was observed. Photograph taken on 13 November 2014.
Figure 5. Scanning electron micrograph showing individual bacterial cells on smooth (A) and frosted (B) surface of glass slides at depth of 5.5 m and clusters of bacterial cells on smooth (C) and frosted (D) surface of slide at depth of 3 m. Black bar denotes 1 μm on images A, B, C, and 10 μm on image D. Images were taken with field-emission scanning electron microscope.
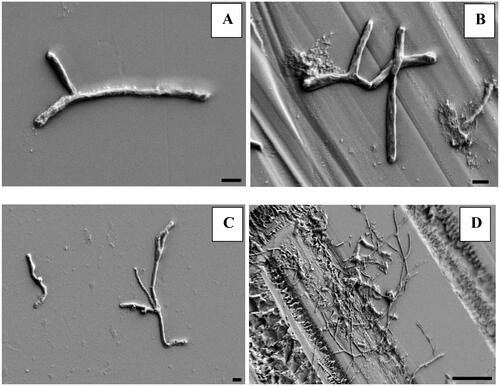
Figure 6. Representation of 1-week (sample 9), 2-week (sample 10) and 4 m depth microbial communities’ diversity (sample 4) by 16S rDNA amplicon sequencing. Only genera with more than 1% abundance are shown and ‘other’ refers to the detected genera with lower abundance (see the interactive supplementary table S3 for full description). See discussion for more info.
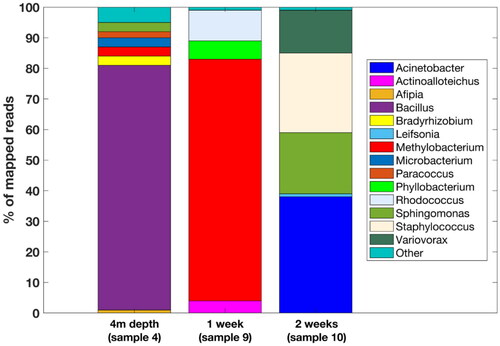
Table 7. Doses per glass slide.