Figures & data
Table 1. A list of Porphyra containing the 1506 ribosomal introns sequenced for phylogenetic and intra-specific analysis.
Table 2. PCR profiles used for group-I intron amplification.
Fig. 1. An agarose gel (representing a subset of 742 Porphyra samples) illustrating an initial amplification screening for the presence of 1506 introns with flanking primers porint1 and porint2. The lanes show variable size introns or templates that lacked intron amplification. All fragments showing the 174 bp band size represent samples lacking the 1506 intron (shown as the top band of a doublet, the lower band represents primer dimer). The first lane (M) is a ΦX/Hae III DNA size marker. Numbers attached to arrows represent amplification size class and actual intron size (parentheses) in bp. Lane 1–2: P. purpurea (Orr's Island, Maine); 3–10: P. linearis (Rachel Carson Salt Pond Reserve, Bristol, Maine); 11–15, 17–19: P. umbilicalis (Peak's Island, Casco Bay, Maine); 16: Negative control; 20: P. linearis (Old Soaker, Mount Desert Island, Maine).
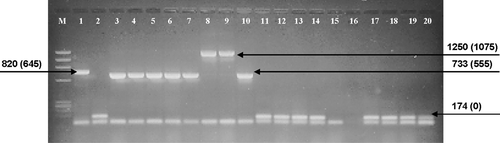
Table 3. Initial screening for the 1506 intron from North Atlantic Porphyra collections using flanking primers porint1 and porint 2.
Table 4. The number of 1506 intron amplifications in a sub-sample (total N = 181) from the Porphyra survey. Table shows the difference in amplifications between using only flanking primers (porint 1 & porint 2) or the combination of a flanking and intron specific primer combination (porint 5 and porint 6).
Table 5. Amplification results of 516 Group-I introns using the Int5pr1 and Int5pr2 (intron specific) primer pair from a survey of 88 Porphyra individuals.
Fig. 2. Similar putative endonuclease sequences from the Bangiophyceae within the 1506 ribosomal group-I intron. The amino acid sequence of the Porphyra umbilicalis intron ORF found in four individuals (Pumb) is aligned to the corresponding sequences from Porphyra spiralis (Psp 1), and Bangia atropurpurea (Bat 1) as given in Haugen et al. (Citation1999). Identical positions are indicated by dots, deletions by dashes. The His-Cys box motif ( bold line) is compared to the homing endonuclease I-PpoI. Conserved residues proposed to be directly involved in zinc binding (C100, C105, H110, C125, C132, H134, C138) and the active site (H98, N119) of the I-PpoI endonuclease (Flick et al., Citation1998) are indicated by shading.

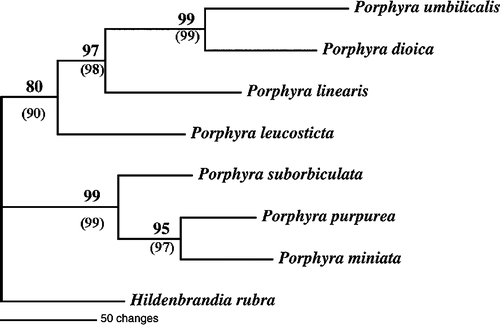
Fig. 3. Phylogram of the 1506 Group-I Intron from seven North Atlantic Porphyra using Maximum Parsimony analysis. Numbers represent bootstrap support from resampling of 1000 replicates. Parentheses = bootstrap support from weighted analysis.