Figures & data
Fig. 1. Schematic drawing of novel lineage (C) arising from within the parental genus B. B′ denotes the sister taxon of B that must go extinct if C is to be considered monophyletic.
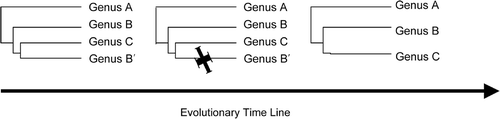
Fig. 2. Variability map of the 18S rRNA from http://bioinformatics.psb.ugent.be/webtools/rRNA/varmaps/Scer_ssu.html
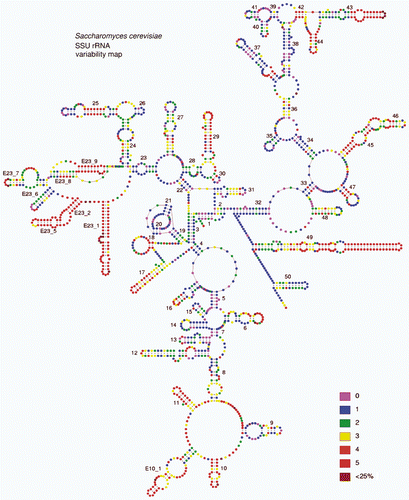
Fig. 3. Secondary structure of the V4 region from (A) Bolidomonas, (B) Stephanopyxis, (C) Rhizosolenia, (D) Porosira, (E) Cymatosira, (F) Ditylum, (G) Fragilaria, (H) Nitzschia and (I) Toxarium. Redrawn with permission from http://www.rna.ccbb.utexas.edu/DAT/3B/Standard/index.php?xysub=1&organism=Toxarium&seq_size=&rna_type=&orf=&rna_class=&from_gene=&structure=ALL&cell_loc=&ord=&xyac_info=m&xybegin=0&xyrange=50&xyco=yes&query_type=results.
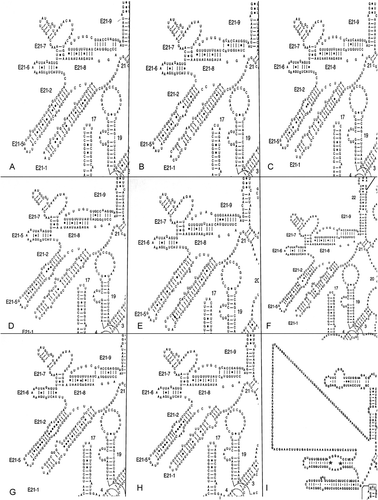
Fig. 4. Alignment of the helices E21 and E22 using CLUSTAL (A) and a secondary structure model (B). Note there is an 18 base-pair shift in the base numbering in the secondary structure model. In the secondary structure alignment, the marks under the bases refer to the type of bond that base has in the helix. Key: ∼: strong Watson and Crick pairing, viz., C–G or A–T; -: weak Watson and Crick pairing, viz., G–A; =: weak Watson and Crick pairing, viz., G–U;.: loop no pairing; +: weak Watson and Crick pairing, viz., U–U; #:alignment breaks the secondary structure, this mark reveals a base in the wrong position and that base must be moved until the mark under the base reflects the best Watson and Crick pairing that can be obtained.

Fig. 5. Phylogenetic trees constructed with a neighbour joining method from the full 18S rRNA gene in alignment (A) and (B).
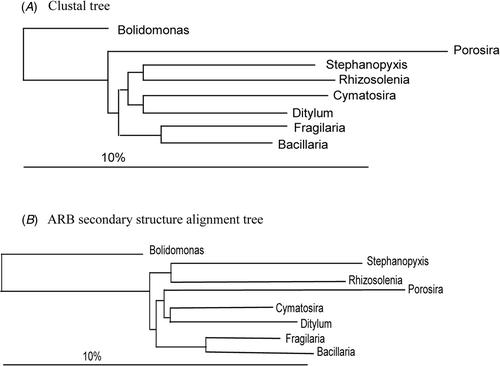
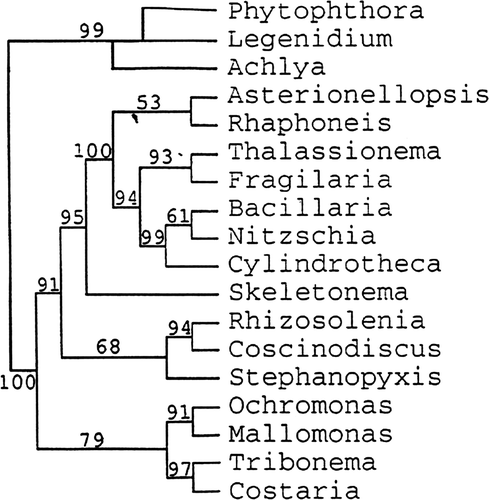
Fig. 6. Clustal alignment trees reproduced from Medlin et al. (Citation1993). Tree made by D. M. Williams.
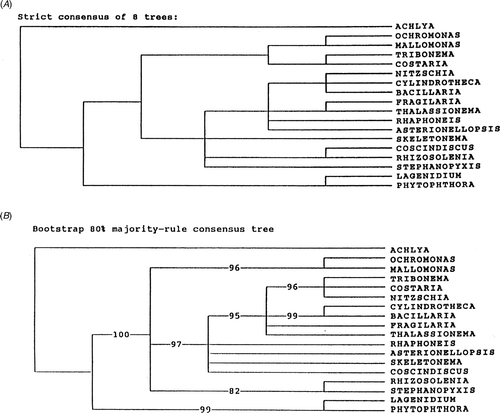
Fig. 7. Secondary structure tree reproduced from Medlin et al. (Citation1993). Tree made by L. K. Medlin.