Figures & data
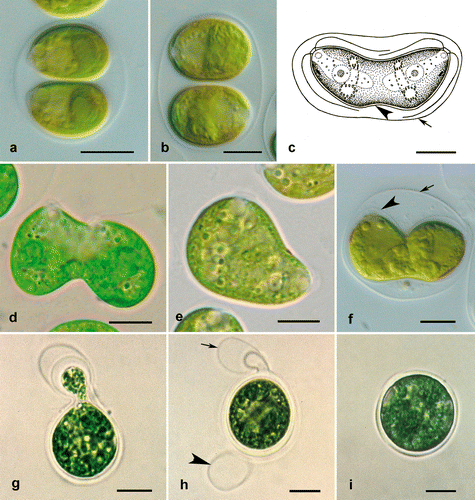
Fig. 1. Molecular phylogeny of the clockwise group of the Chlorophyceae, based on SSU rDNA sequence comparisons. The phylogenetic tree shown was inferred by maximum likelihood method based on a dataset of 1524 aligned positions of 85 taxa using PAUP 4.0b10. For the analysis, the GTR model (base frequencies: A 0.2465, C 0.2107, G 0.2819, T 0.2609; rate matrix: A-C 1.1409, A-G 2.6738, A-T 1.4865, C-G 0.9087, C-T 5.3353, G-T 1.0000) with the proportion of invariable sites (I = 0.5213) and gamma distribution shape parameter (G = 0.5923) was chosen, which was calculated as best model by Modeltest 3.7. Bayesian values (>0.95) were calculated by MrBayes 3.1 (first value in boxes) using the covarion settings (5 million generations) and PHASE 2.0 (second value) using THREESTATE and RNA7D models for unpaired and paired nucleotides respectively. Bootstrap values (>50%) of the maximum likelihood (using the GTR + I + G model, 100 replicates; third value) using PAUP, the randomized accelerated maximum likelihood using the RAxML 7.0.3 (using the GTR + I + G model, 1000 replicates; fourth value), neighbour-joining (using the GTR + I + G model, 100 replicates; fifth value), and maximum parsimony (1000 replicates; sixth value) were marked in boxes and only given for the clades. The strains marked in bold are new sequences in this study. Strain and accession numbers are given after the species name. The clade designation follows Pröschold & Leliaert (2007). The Chaetophora-clade was used as outgroup.
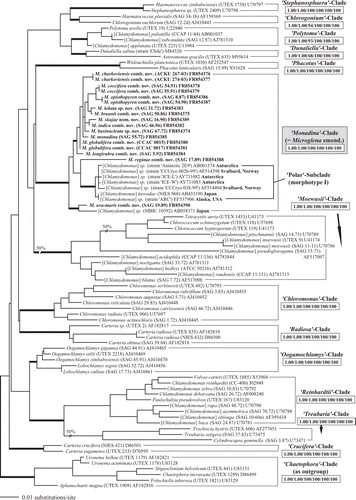
Fig. 2. Comparison of the conserved DNA Barcode region of ITS-2 among species of Microglena. The upper part of the figure shows the ITS-2 secondary structure of SAG 55.72 M. monadina, including details of the stem of 5.8S and LSU rDNA, and helices I–III. The conserved regions are highlighted in grey boxes. The bottom part of the figure shows the conserved region of ITS-2 aligned manually or automatically using MARNA. Helices I-IV of ITS-2 are drawn with straight lines for practical reasons.
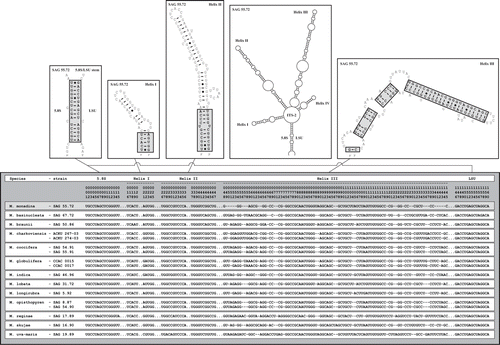
Fig. 3. ITS-2 DNA Barcode of the Microglena species. The sequence alignment presented in was translated into a base-pair alignment using a number coding for each base-pair. The base positions in the sequence alignment are given for comparison. The barcode for each species is named by a letter (A–M). Unique compensatory base changes and non-homoplasious synapomorphies for Microglena species are marked by asterisks.
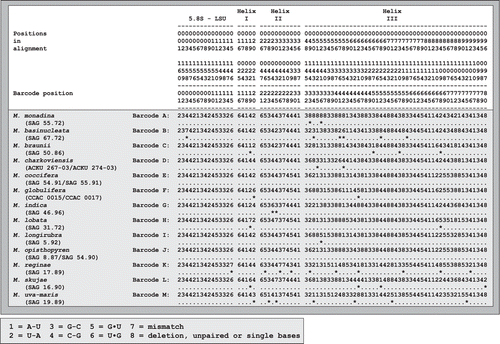
Fig. 4. Molecular phylogeny of the ITS-2 DNA Barcode in comparison to the morphotypes in Microglena. The phylogenetic tree represents a majority-rule consensus of five equal trees (120 steps) calculated by maximum parsimony in PAUP. The asterisk marks the only branch that varies among the five trees. Morphotypes I–III are characterized by chloroplast shape and the shape and position of the pyrenoid. Each species is documented by a line drawing and grouped according to morphotype.
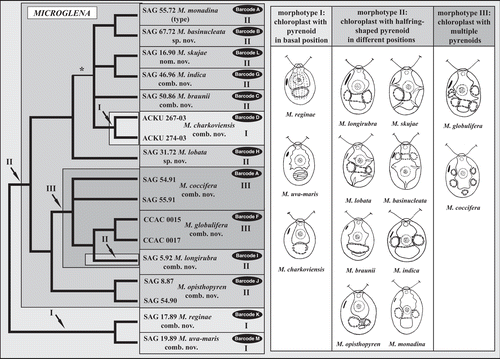
Table 1. Compensatory base changes (CBCs) and uncorrected p-distances among the ITS-2 DNA barcodes of the Microglena species. The upper-right half of the table shows the total number of compensatory changes (CBC/Hemi-CBCs), while the lower-left half gives the uncorrected p-distances calculated in PAUP.
Fig. 5. Morphology of vegetative cells in Microglena. a–d. Microglena monadina (SAG 55.72). e–h. Microglena braunii (SAG 50.86). i–l. Microglena coccifera (SAG 55.91). m–p. Microglena charkoviensis (ACKU 267-03). a, b, d–j and l–p show general views of vegetative cells; c, sporangium, k, cell in apical view (optical section). Scale bars = 10 µm.
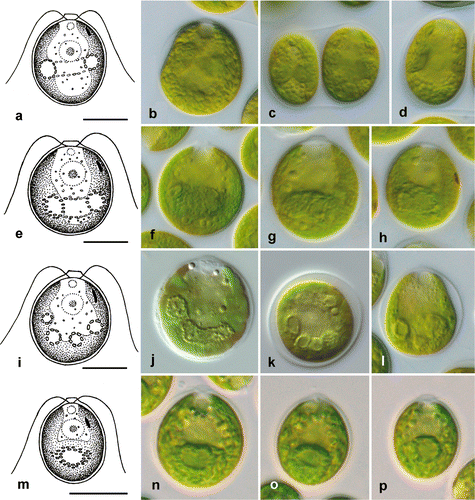
Fig. 6. Morphology of vegetative cells in Microglena. a–d. Microglena globulifera (CCAC 0017). e–h. Microglena indica (SAG 46.96). i–l. Microglena skujae (SAG 16.90). m–p. Microglena opisthopyren (SAG 8.87). a–j and l–p show general views of vegetative cells; k, sporangium. Scale bars = 10 µm.
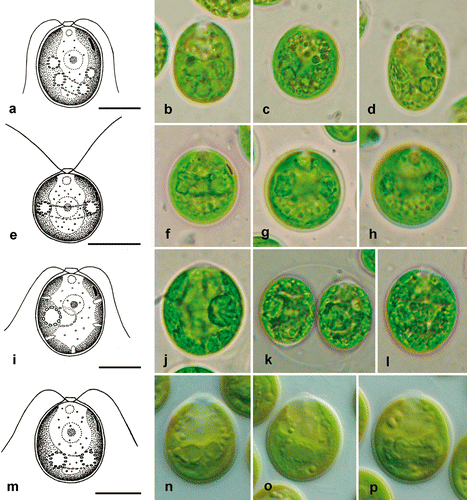
Fig. 7. Morphology of vegetative cells in Microglena. a–d. Microglena uva-maris (SAG 19.89). e–i. Microglena longirubra (SAG 5.92). j–m. Microglena lobata (SAG 31.72). n–q. Microglena basinucleata (SAG 67.72; arrows indicate the position of the pyrenoid, which is unclear). a–f, j, k and n–q show general views of vegetative cells; g–i, l and m, surface views. Scale bars = 10 µm.
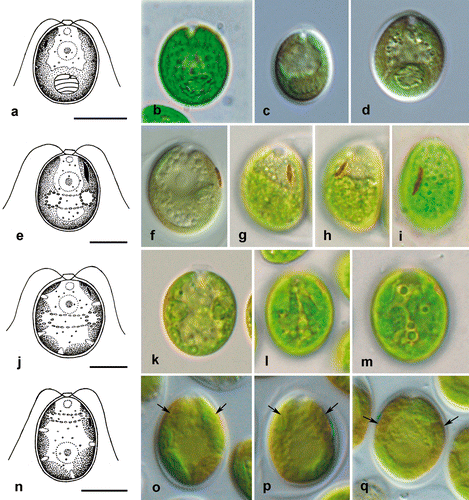
Table 2. Morphological characters and reproduction of investigated species.
Fig. 8. Reproduction in Microglena. a, b. Asexual reproduction by zoosporulation (a without rotation of protoplast, b with rotation of protoplast by 90°). c–f. Incomplete cell division similar to asexual reproduction by protocytotomy; note the extended mother cell wall (arrows) and newly synthesized daughter cell walls (arrowheads). g–i. Sexual reproduction by advanced anisogamy: g shows fusing macro- and microgametes, h an empty microgametangium (arrow) and an empty macrogametangium (arrowhead) on the wall of the young zygote, and i a young zygote. a Microglena opisthopyren (SAG 8.87), b, d Microglena longirubra (SAG 5.92), c Microglena monadina (SAG 55.72), e Microglena lobata (SAG 31.72), f–i Microglena coccifera (SAG 55.91). Scale bars = 10 µm.