Figures & data
Fig. 1. Phylogenetic tree of Cyclotella species and the strains used to investigate developmental plasticity based on Bayesian analysis (TN + G) of partial 28S rDNA sequences (D1/D2 regions). The numbers at the nodes are posterior probabilities (PP) above 0.50. DNA sequences determined in this study are in bold. Scale bar represents 3 substitutions per 100 nucleotides.
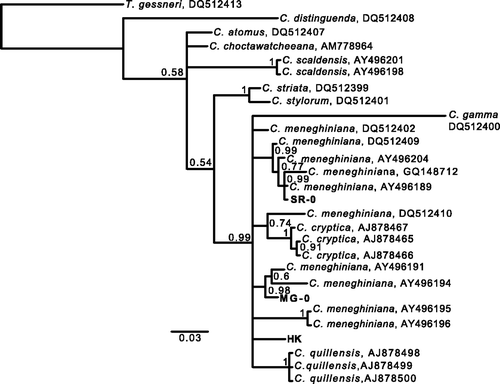
Table 1. Strain characteristics.
Fig. 2. Variables in the morphological analyses, illustrated on a C. meneghiniana valve. A, number of CFP (central fultoportulae); B, number of striae; C, valve diameter; and D, diameter of the central area.
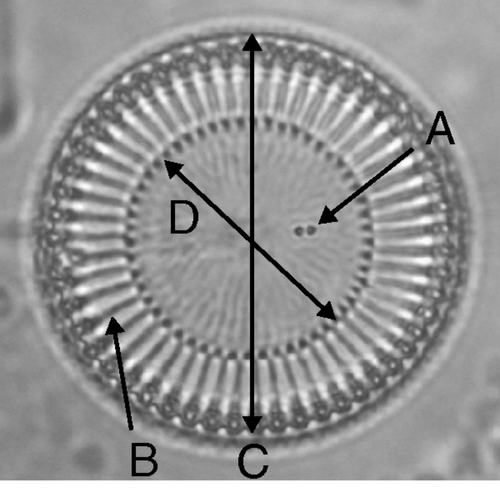
Table 2. Collection sites and water quality data.
Fig. 3. Morphological traits of seven clonal strains of C. meneghiniana in freshwater medium (FW: salinity 0) and 50% seawater medium (SW: salinity 17 psu). Reaction norms are shown for (a) the number of central fultoportulae (CFP), (b) the number of striae, (c) the diameter of the central area, and (d) the valve diameter. Bars indicate mean ± standard error (SE).
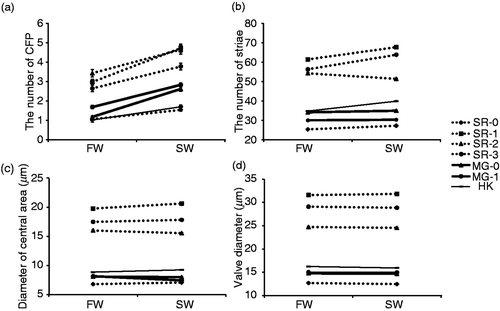
Table 3. The results of GLM summarizing the effect of salinity and strain on the traits of C. meneghiniana; d.f. = degrees of freedom.
Table 4. Pairwise comparison of the effect of salinity in each strain. ‘Increase’ and ‘decrease’ tabulate significant changes (P < 0.05) in seawater vs. freshwater; n.s. = not significant.
Table 5. Pairwise comparison of G × E interactions (SR-0, MG-0 and HK).
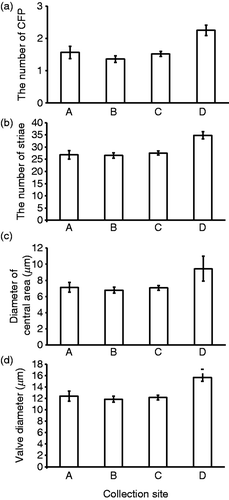
Fig. 4. Morphological traits of specimens at each collection site (A, B, C and D). (a) Number of central fultoportulae (CFP), (b) number of striae, (c) diameter of the central area, (d) valve diameter. Bars indicate mean ± standard error (SE).