Figures & data
Table 1. The 42 out of 166 isolates from Fucus vesiculosus (F) and Delesseria sanguinea (D) that could be affiliated to 17 type strains previously described as associated with algae by other authors (see References). The similarity (%) to the closest type strain and the GenBank accession number are given, and the number of isolates.
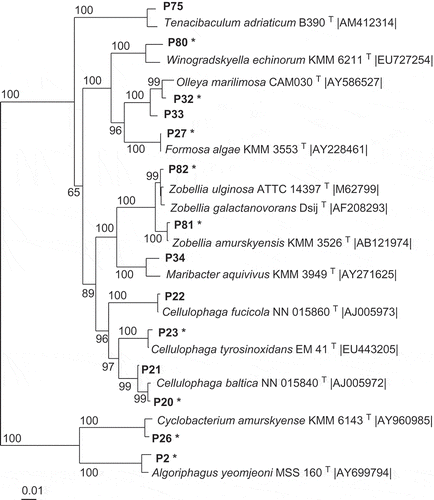
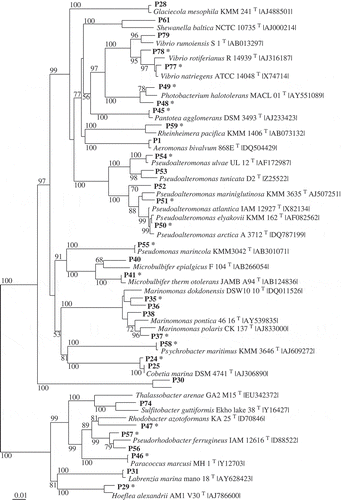
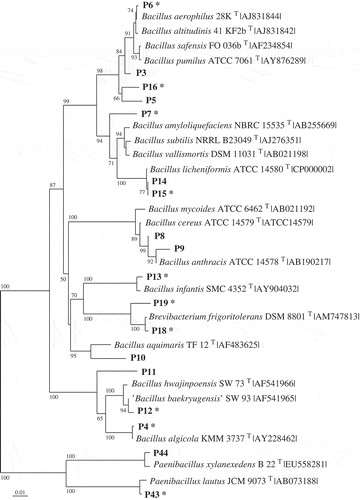
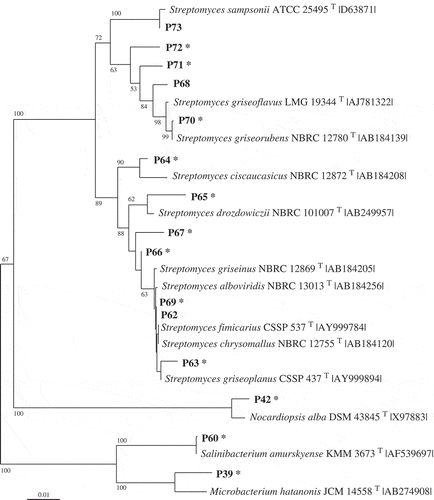
Table 2. Phylotypes (Px) associated exclusively with Fucus vesiculosus or Delesseria sanguinea, with the closest type strain (in parentheses: for the strain number see Supplementary Information 1). The phylotypes affiliate to seven classes of bacteria according to the 16S rRNA gene sequences. S, phylotype active against the standard set; M, phylotype against macroalga-associated set; – not active; nd, not determined.
Table 3. Total number and percentage of phylotypes with antibiotic activity, classified according to their origin, viz. Fucus vesiculosus (a), Delesseria sanguinea (b), and strains shared by both macroalgae (c). In each case information is given about the number of strains that are active against standard test strains (‘Standard set’: Bacillus subtilis, Staphylococcus lentus, Escherichia coli and Candida glabrata), and against the macroalga-associated strains (‘Macroalga-associated set’), the latter being reported as totals and also as ‘surface-associated strains’ (Bacillus algicola, Formosa algae and Algicola bacteriolytica) and ‘macroalgal pathogen strains’ (only Pseudoalteromonas elyakovii). The total active phylotypes represent the number of different phylotypes by algae that inhibited at least one test strain.
c. Shared by both macroalgae