Figures & data
Table 1. Morphology type, collection information, GenBank accession numbers and previous papers for rbcL and cox1 sequences of Compsopogon specimens used in this study. New sequences in bold
Table 2. Percent difference (lower left matrix) and base pair changes (upper right matrix) among the seven unique rbcL haplotypes for Compsopogon specimens. Location codes as in
Table 3. Percent difference (lower left matrix) and base pair changes (upper right matrix) among the seven unique cox1 barcode haplotypes for Compsopogon specimens. Codes as in
Table 4. Physical and chemical parameters of collection locations. Codes as in
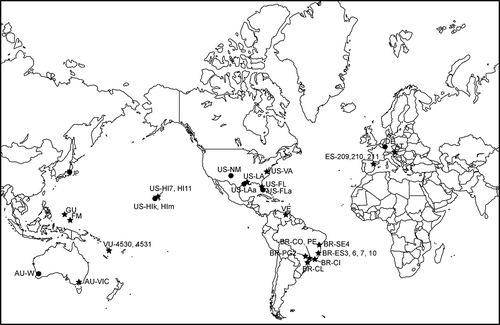
Figs 1. World map showing locations of the new specimens sequenced for this study (stars) and previously sequenced specimens from GenBank (circles). Location codes as in .
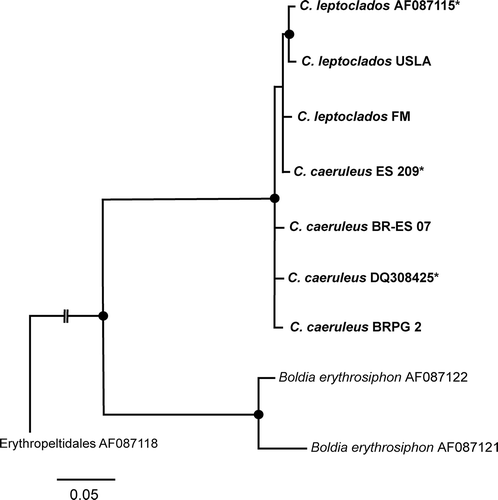
Figs 2. Maximum likelihood (ML) analysis tree (-Ln 11094.402668) showing the relationships within the Compsopogonales (with representatives of Compsopogon and Boldia) based on rbcL gene data, with the Erythropeltidales as the outgroup. Nodes with filled circles had statistical support ≥ 0.90 Bayesian posterior probability and 90 ML bootstrap values. Branches without values had statistical support < 0.70 posterior probability and 70 ML bootstrap. Haplotypes marked with * represent more than one sequence. Haplotypes AQUA and US-HI7 correspond to AF087115 and DQ308425, respectively: see and for information on haplotypes.
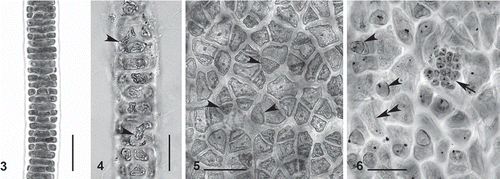
Figs 3–6. Compsopogon caeruleus: photomicrographs of important morphological features for both the ‘caeruleus’ and ‘leptoclados’ morphologies. Location codes as in Table 1. 3. Early cortication of ‘caeruleus’ morphology showing regular cortication. Specimen from BR-ES7. 4. Early cortication of ‘leptoclados’ morphology showing irregular cortication and rhizoidal cells (arrow-heads). Specimen from BR-CL. 5. Surface view of ‘caeruleus’ morphology showing regular (polygonal) cortical cells and monosporangia (arrowheads). Specimen from BR-ES3. 6. Surface view of ‘leptoclados’ morphology showing monosporangia (arrowheads), microsporangia (arrow) and rhizoidal cell (double arrowhead). Specimen from BR-CL. Scale bars = 50μm.