Figures & data
Figs 1. Colonization of the algae on the royal palm (Roystonea regia). 1. Palms with algae growing vigorously on the trunk in a busy street in Haikou, Hainan Province, China. 2. Palms with poor algal growth in the Xinglong Tropical Botanical Garden, CATAS, Wanning County, Hainan. 3. Close-up, showing algae on the bark. 4. Bark section showing algae on the surface.

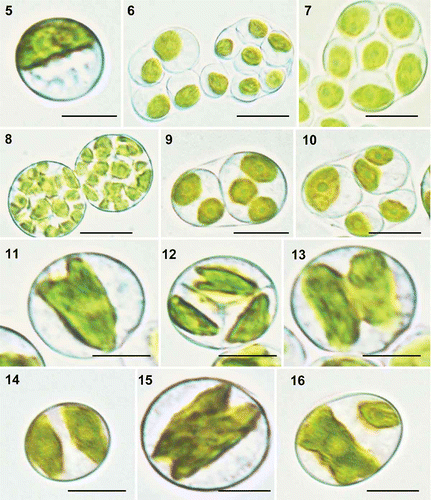
Figs 2. Heveochlorella roystonensis, strain ITBB A3-8, LM. 5. Mature vegetative cell with a parietal cup-shaped chloroplast. 6. Sporangia with four or eight autospores. 7. Sporangium with eight autospores. 8. Sporangia with 32 autospores. 9. Sporangium with two autospores, each containing two or three chloroplasts. 10. Sporangium with four autospores of different sizes. 11–13. Dividing chloroplasts. 14. Cell after completion of chloroplast division. 15. Chloroplast dividing from both ends. 16. Unequal division of a chloroplast. Scale bar = 5 μm.
Figs 3. Heveochlorella roystonensis, strain ITBB A3-8, TEM. 17, 18. Mature vegetative cell showing the cup-shaped parietal chloroplast, pyrenoid, nucleus and mitochondria. 19. Two-layered cell wall of mature cell. 20. Sporangium containing multiple autospores. 21, 22. Ultrastructure of pyrenoids showing tube-like invaginations arranged radially. C, chloroplast; M, mitochondria; N, nucleus; V, vacuole; CW, cell wall. Scale bars = 1 μm.
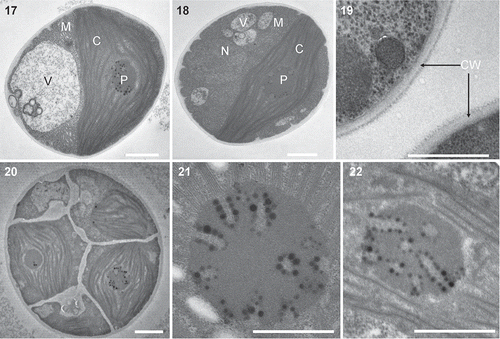
Table 1. Morphological comparison between Heveochlorella roystonensis ITBB A3-8 and three closely related strains, Heveochlorella hainangensis FGG01, Kalinella bambusicola CAUP H7901 and Heterochlorella luteoviridis SAG 211-2a
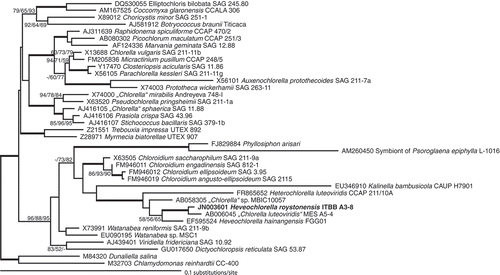
Figs 4. Phylogenetic tree deduced from nuclear 18S rDNA sequences of Heveochlorella roystonensis and related trebouxiophycean green algae. The tree was rooted with two chlorophycean sequences as outgroups. Tree topology is based on a maximum likelihood (ML) analysis, which was mostly consistent with trees calculated by maximum parsimony (MP) and neighbour joining (NJ). The numbers at nodes indicate bootstrap support for NJ/MP/ML (see Methods). Statistical support of more than 90% for all three methods is indicated by a thick line. Bootstrap values lower than 50% are not shown. Branch lengths reflect the evolutionary distances indicated by the scale. GenBank accession numbers precede the taxon names.
Figs 5. Phylogenetic tree based on concatenated 18S+ITS sequence data of Heveochlorella and Heterochlorella with two more remotely related trebouxiophycean green algae as outgroups. A maximum likelihood analysis was used to infer the tree topology. The bootstrap support for NJ/MP/ML is indicated at the nodes (see Methods). Branch lengths reflect the evolutionary distances indicated by the scale. GenBank accession numbers precede the taxon names.

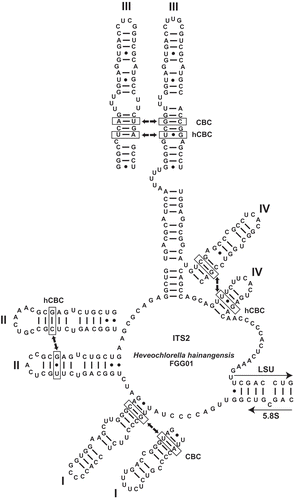
Figs 6. Secondary structure model of the internal transcribed spacer 2 (ITS2) of Heveochlorella hainangensis FGG01. Helices I, II, IV and the conserved part of helix III are shown for H. roystonensis ITBB A3-8 for comparison. Hemi-compensatory base changes (hCBCs) and full compensatory base changes (CBCs) are indicated for each helix.