Figures & data
Table 1. Origins of the cyanobacterial isolates investigated and their possible assignment as botanical species on the basis of morphology, morphometry and ecology.
Fig. 1. Percentages of total phycobiliproteins in different Scytonema cultures. Scytonema isolate codes are given in .
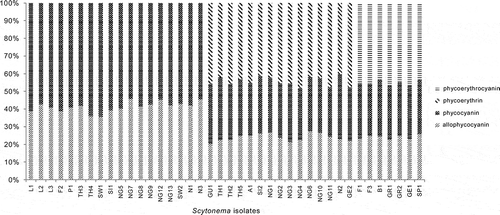
Fig. 2. Cluster analysis of Scytonema isolates according to their pigment characteristics. Scytonema isolate codes are given in .
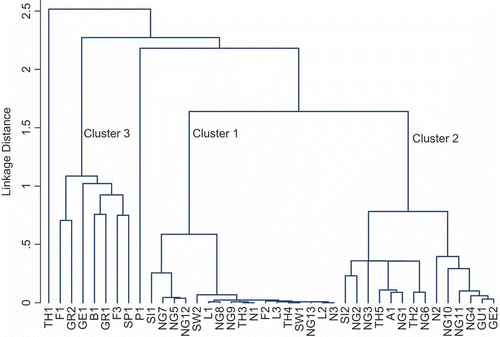
Table 2. Characteristics of the three groups of Scytonema distinguished by cluster analysis of biochemical data ().
Table 3. Comparison of the phycobiliproteins, scytonemin, carotenoids and mycosporine-like amino acid data (mean ± S.D.), evaluated by non-parametric Kruskal–Wallis tests (P < 0.05) among the three groups of Scytonema cultures distinguished by cluster analysis ().
Table 4. Mycosporine-like amino acid compounds and scytonemin in different isolates of Scytonema. The numbers represent the order of leaving the HPLC column. The specific MAA contents are expressed as absorbance (at 335 nm) mg−1 chl a. The specific scytonemin contents are expressed as absorbance (at 370 nm) mg−1 chl a. – = not detected. For Scytonema isolate codes, see .
Fig. 3. Percentages of total carotenoids in different Scytonema cultures. Scytonema isolate codes are given in .
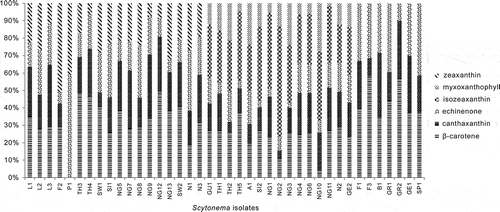
Table 5. Comparison of the phycobiliproteins, scytonemin, carotenoids and mycosporine-like amino acid data among the different habitat groups of Scytonema, evaluated by non-parametric Kruskal–Wallis tests (P < 0.05). The habitats represented among the 40 isolates were aerophytic (2), epilithic (7), cavernicolous (2), atmophytic (18), edaphic (4), and corticolous (7).