Figures & data
Fig. 1. Summary of the nomenclatural problems in the case of some coccoid green algal isolates [Chlorella minutissima Lefevre ALCP no. 87, ‘C. minutissima’ C 1.1.9 and SAG 1.80, Nannochloris eucaryotum Mainz 1 and ‘N eucaryotum’ UTEX 2502 (=SAG 55.87)] according to Fott & Nováková (Citation1969), Andreyeva (Citation1975), Wilhelm et al. (Citation1982), Kalina & Puncochárová (Citation1987), Menzel & Wild (Citation1989), Huss et al. (Citation1999), Krienitz et al. (Citation1999), Yamamoto et al. (Citation2003) and Henley et al. (Citation2004). Misinterpreted strains (C 1.1.9; SAG 1.80 and UTEX 2502) are framed with thin dashed lines and authentic strains with thick, continuous lines. Strains possessing nearly identical 18 rRNA gene sequences are grouped in a box with thick dashed lines and are the subjects of this study.
![Fig. 1. Summary of the nomenclatural problems in the case of some coccoid green algal isolates [Chlorella minutissima Lefevre ALCP no. 87, ‘C. minutissima’ C 1.1.9 and SAG 1.80, Nannochloris eucaryotum Mainz 1 and ‘N eucaryotum’ UTEX 2502 (=SAG 55.87)] according to Fott & Nováková (Citation1969), Andreyeva (Citation1975), Wilhelm et al. (Citation1982), Kalina & Puncochárová (Citation1987), Menzel & Wild (Citation1989), Huss et al. (Citation1999), Krienitz et al. (Citation1999), Yamamoto et al. (Citation2003) and Henley et al. (Citation2004). Misinterpreted strains (C 1.1.9; SAG 1.80 and UTEX 2502) are framed with thin dashed lines and authentic strains with thick, continuous lines. Strains possessing nearly identical 18 rRNA gene sequences are grouped in a box with thick dashed lines and are the subjects of this study.](/cms/asset/b79c7266-3253-4d5e-94cf-3237b18c0735/tejp_a_854411_f0001_b.gif)
Table 1. Oligonucleotide primers used in this study.
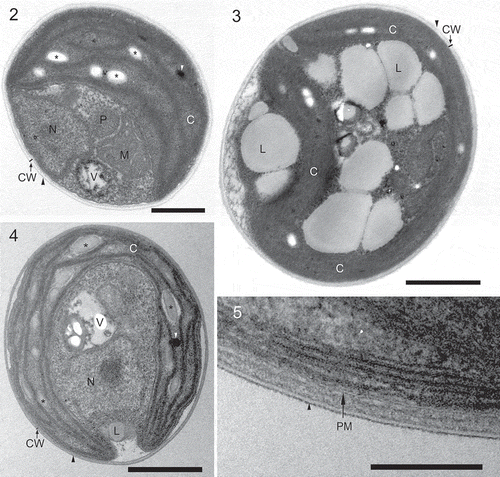
Figs 2–5. Pseudochloris wilhelmii, transmission electron micrographs of SAG 55.87. C, chloroplast; CW, cell wall; L, lipid droplet; M, mitochondrion; N, nucleus; P, peroxisome; PM, plasma membrane; V, vacuole. Black and white arrowheads indicate the outer trilaminar layer of the cell wall and plastoglobuli, respectively. Asterisks indicate starch grains. 2, 4. Vegetative cell. 3. Vegetative cell with lipid droplets. 5. A close-up from showing the cell wall structure: note the outer trilaminar layer and the granulo-fibrillar inner layer covering the plasma membrane. Scale bars = 0.5 µm (), 1 µm (, ), and 0.2 μm ().
Figs 6–11. Pseudochloris wilhelmii, transmission electron micrographs of SAG 55.87 during autosporulation. C, chloroplast; DCW, daughter cell wall; M, mitochondrion; MCW, mother cell wall; PM, plasma membrane; T, thylakoid; V, vacuole. Single and double black arrowheads indicate the outer trilaminar layer of the cell wall of the daughter cell and of the mother cell, respectively. Asterisks indicate starch grains, white arrowheads show plastoglobuli. C1, C2, C3 and C4 correspond to the chloroplasts of the different daughter cells within the sporangium. 6. Mother cell with two autospores. 7. Mother cell with four autospores. 8. A daughter cell at a late phase of autosporulation. 9. A close-up from , showing the structure of the mother cell wall and the developing daughter cell walls. The daughter cell wall has a fine trilaminar layer and a firm granulo-fibrillar inner layer. 10. A close-up from , showing the structure of the mother cell wall and the developing daughter cell walls. At this stage, the daughter cell wall has only a fine trilaminar layer. 11. A close-up from , showing the rupture of the mother cell wall. Scale bars = 1 µm (–) and 0.2 μm (–).
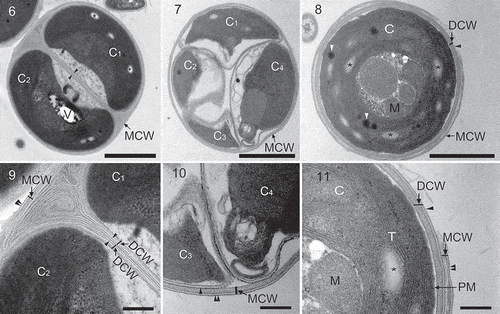
Fig. 12. HPLC chromatogram of pigments extracted from SAG 55.87 at 440 nm. Retention time (min) on the x-axis and absorbance (mAU) on the y-axis. Peak identities: (1) neoxanthin, (2) violaxanthin, (3) antheraxanthin, (4) lutein, (5) zeaxanthin, (6) chlorophyll b, (6a) isomer of chlorophyll b, (7) chlorophyll a, (7a) isomer of chlorophyll a, (8) α-carotene and (9) β-carotene.
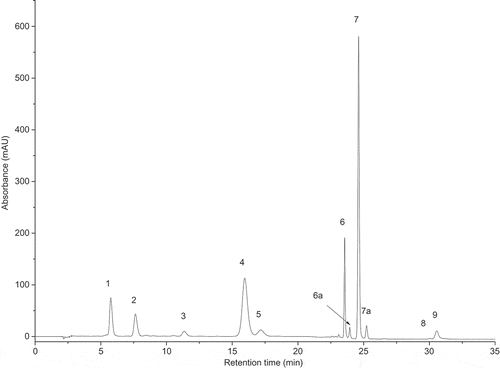
Table 2. Isolated pigments of SAG 55.87 in relation to the concentration of chlorophyll a: measurements are mean ± s.d. (N = 3).
Fig. 13. Maximum likelihood (ML) tree of 18S rRNA gene sequences retrieved from the studied isolates (SAG 55.87, SAG 1.80 and UTEX 2502) and their closest relatives (Trebouxiophyceae, Chlorophyta) based on 1691 nucleotide positions (best tree likelihood score, lnL = –4575.832). Bootstrap values greater than 70 [based on 100 and 1000 replicates for ML and maximum parsimony (MP), respectively] and Bayesian (B) posterior probabilities higher than 0.70 are shown (ML/MP/B). Sequences determined in this study appear in bold. The number of introns is shown in square brackets, if present. Asterisks mark authentic strains according to Henley et al. (Citation2004), Luo et al. (Citation2010), Krienitz et al. (Citation2011), Somogyi et al. (Citation2011) and this study.
![Fig. 13. Maximum likelihood (ML) tree of 18S rRNA gene sequences retrieved from the studied isolates (SAG 55.87, SAG 1.80 and UTEX 2502) and their closest relatives (Trebouxiophyceae, Chlorophyta) based on 1691 nucleotide positions (best tree likelihood score, lnL = –4575.832). Bootstrap values greater than 70 [based on 100 and 1000 replicates for ML and maximum parsimony (MP), respectively] and Bayesian (B) posterior probabilities higher than 0.70 are shown (ML/MP/B). Sequences determined in this study appear in bold. The number of introns is shown in square brackets, if present. Asterisks mark authentic strains according to Henley et al. (Citation2004), Luo et al. (Citation2010), Krienitz et al. (Citation2011), Somogyi et al. (Citation2011) and this study.](/cms/asset/da8628c7-1a1a-4daa-9bd7-f0deaa391ea5/tejp_a_854411_f0005_b.gif)