Figures & data
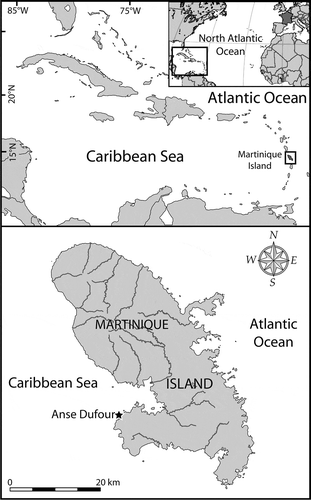
Fig. 1. Maps showing the location of Martinique Island in the Caribbean Sea (Atlantic Ocean), and the sampling area (Anse Dufour) on the western coast of the island.
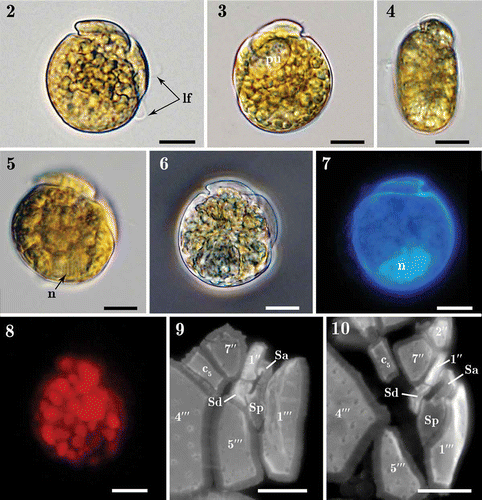
Figs 2–10. Light micrographs of Madanidinium loirii. Fig. 2. Right lateral view of a living cell with the longitudinal flagellum (lf) visible. Fig. 3. Left lateral view of a living cell with a pusule (pu) visible. Fig. 4. Dorsal view of a living cell showing the epitheca inclined toward the right side. Fig. 5. Right view of a cell with focus on the nucleus (n). Fig. 6. Right lateral view of a fixed environmental specimen used for single-cell molecular analysis (isolate IFR 12–200). Fig. 7. Left lateral view of a DAPI-stained specimen showing the posterior position of the nucleus (n). Fig. 8. Right lateral view of a living cell seen in epifluorescence (blue excitation) showing chlorophyll autofluorescence and the presence of small discoid chloroplasts. Figs 9–10. Detail of sulcal plates of two specimens stained with Calcofluor white. Except in Fig. 6, all specimens are from strain IFR–MLO–02M. Scale bars: 10 µm.
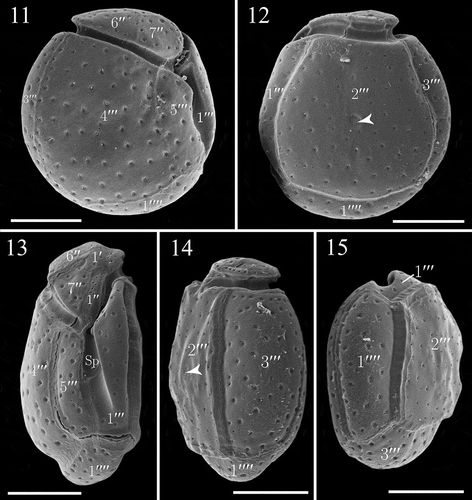
Figs. 11–15. SEM micrographs of Madanidinium loirii from strain IFR-MLO-02M. Fig. 11. Right lateral view (holotype specimen). Fig. 12. Left lateral view, note the reduced epitheca and area of densely arranged pores (arrowhead). Fig. 13. Ventral view showing the tilted epitheca. Fig. 14. Dorso-lateral view (arrowhead pointing to the area of densely arranged pores). Fig. 15. Antapical view. Scale bars: 10 µm.
Figs. 16–23. Details of the theca of Madanidinium loirii in SEM. Fig. 16. Apical view. Fig. 17. Right lateral side of the epitheca. Fig. 18. Left lateral side of the epitheca. Fig. 19. Ventral view of the epitheca, note the flagellar pore (fp) visible partially. Fig. 20. Dorsal view. Fig. 21. Detail of thecal surface with groups of pores and some isolated pores. Fig. 22. Oval area of densely arranged pores on the 2′′′ plate. Fig. 23. Area of pores on the 2′′′ plate of another specimen, note that the shape is elongated. Scale bars: 5 µm in Figs 16–20; 1 µm in Figs 21–23.

Figs. 24–28. Line drawings of Madanidinium loirii. Fig. 24. Representation of a live cell in right lateral view (n: nucleus, pu: pusule). Fig. 25. Ventral view of the theca. Fig. 26. Apical view. Fig. 27. Right lateral view. Fig. 28. Left lateral view. Scale bars: 10 µm in Figs 24, 25, 27, 28 and 5 µm in Fig. 26.
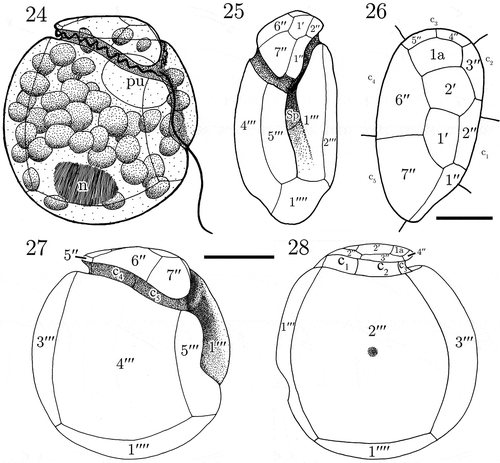
Fig. 29. Maximum likelihood (ML) phylogenetic tree inferred from SSU rDNA (matrix of 77 sequences and 1691 aligned positions). The tree was rooted using Perkinsus marinus sequence as outgroup. Model selected GTR + I + Γ4. Log likelihood = −19792.7. Substitution rate matrix: A ↔ C = 1.52090, A ↔ G = 4.15185, A ↔ T = 1.43273, C ↔ G = 0.81766, C ↔ T = 9.38294, against G ↔ T = 1.00000. Assumed nucleotide frequencies: f(A) = 0.24690, f(C) = 0.19272, f(G) = 0.25795, f(T) = 0.30243. Among site rate variation: assumed proportion of invariable sites I = 0.317. Rates at variable site assumed to be gamma distributed with shape parameter α = 0.511. Bootstrap values (1000 pseudoreplicates) > 65 (in ML) and posterior probabilities > 0.5 (in BI) are shown at nodes, thick lines indicate full support of the branch (100/1.00). ‘+’ indicate nodes present but unsupported. Asterisks indicate benthic taxa with a lateral compression related to M. loirii by morphology.
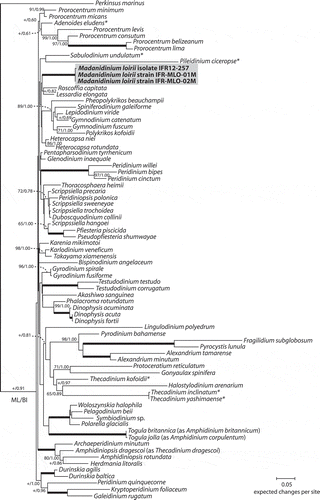
Fig. 30. Maximum likelihood (ML) phylogenetic tree inferred from partial LSU rDNA (matrix of 49 sequences and 860 aligned positions). The tree was rooted using Perkinsus marinus sequence as outgroup. Model selected TN93 + Γ4. Log likelihood = 14250.35343. Transition/transversion ratio for purines = 2.860; transition/transversion ratio for pyrimidines = 7.812. Nucleotides frequencies f(A) = 0.23690, f(C) = 0.18977, f(G) = 0.28854, f(T) = 0.28479. Rates at variable site assumed to be gamma distributed with shape parameter α = 0.528. Only bootstrap values (1000 pseudoreplicates) > 65 (in ML) and posterior probabilities > 0.5 (in BI) are shown at nodes; thick lines indicate full support of the branch (100/1.00); ‘+’ indicates a node present but unsupported. Benthic taxa with a lateral compression are highlighted with asterisks.
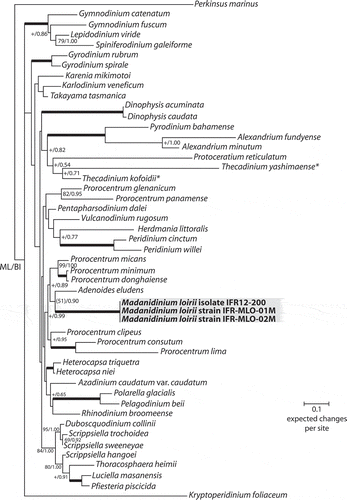
Table 1. Comparative features of Madanidinium loirii and other selected sand-dwelling dinoflagellate genera with a laterally compressed body (Thecadinium excluded), and the planktonic genera Thecadiniopsis and Pseudothecadinium.
