Figures & data
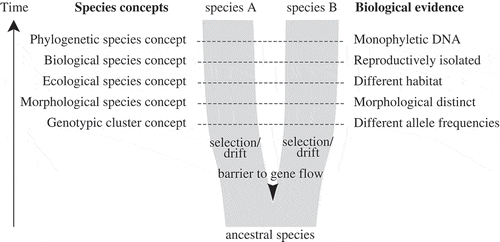
Fig. 1. Simplified diagram of speciation, species concepts and corresponding biological properties of species (after de Queiroz, Citation2007). As populations separate by a barrier to gene flow, independently acting selection and drift will result in two daughter lineages with separate evolutionary trajectories. Through time, these daughter lineages will acquire different properties, which have traditionally served as biological evidence for species delimitation, corresponding to different species concepts. During the process of speciation, these secondary properties do not necessarily arise at the same time or in a regular order, and therefore different species concepts may come into conflict, especially during early stages of speciation.
Figs 2-4. Illustrations of a Wright–Fisher process (a simplified version of the coalescence process), based on Degnan & Rosenberg (Citation2009) and Mailund (Citation2009). Fig. 2. Diagram showing a set of non-overlapping generations where each new generation is sampled from the previous at random with replacement. Each dot represents an individual gene copy and each line connects a gene copy to its ancestor in the previous generation. Starting with a set of individuals in a first generation, the second generation is created by randomly selecting a parent from the first population; the third generation is sampled from the second, and so on. During speciation events (T1 and T2), where populations become separated by a barrier to gene flow, gene copies will only be sampled from within the same population. This coalescence process, running inside the species tree, has two important consequences (Figs 3 and 4). Fig. 3. DNA divergence times may be much older than speciation times, known as ‘deep coalescence’. As illustrated, the most recent common ancestor (MRCA) of two gene copy samples from species A, B and C is much older than the speciation event T1. Fig. 4. Deep coalescence in combination with incomplete fixation of gene lineages within species lineages (incomplete lineage sorting) may result in a gene tree and species tree with different topologies. For example, the gene tree based on three randomly sampled alleles would wrongly suggest a sister relationship between species A and B.
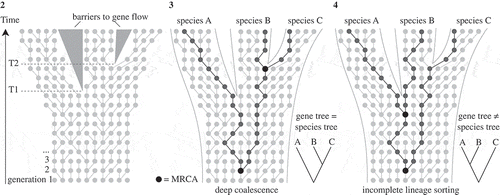
Fig. 5. Neutral coalescence process running within a species tree, based on Klein et al. (Citation1998), Degnan & Rosenberg (Citation2009) and Mailund (Citation2009). Each dot represents an individual gene copy, each colour a different allele, and each line connects a gene copy to its ancestor in the previous generation. Within a population, selection and/or drift will result in changing allele frequencies over time. In the initial stages of lineage splitting, sister species will largely share identical alleles, which has important consequences for species delimitation. In this example, constructing a gene tree at an early stage of speciation would result in none of the three species being monophyletic. Only after sufficient time has gone by, will alleles be completely sorted in each lineage, resulting in reciprocal monophyly for the each of the three species.
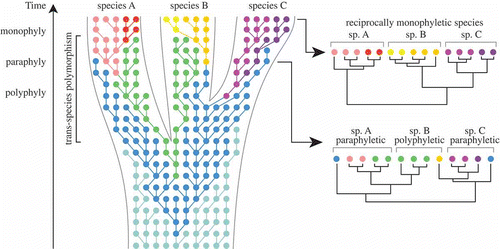
Table 1. A non-exhaustive list of algorithmic species delimitation methods and software packages.
Table 2. Main markers currently employed for DNA-based species delimitation and/or barcoding in some of the principal algal groups. Recommended barcode markers are indicated in bold, although it should be noted that for none of the groups has a consensus been reached.
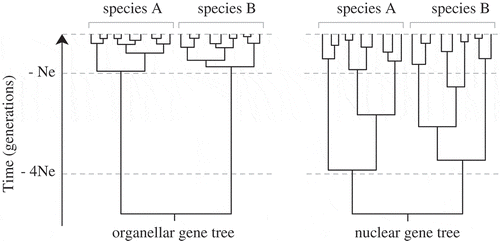
Fig. 6. Expected shapes of nuclear and organellar gene genealogies. Allelic coalescence of a neutral marker is expected to be about four times faster for organellar loci than for nuclear loci, resulting in a shorter time to arise at reciprocal monophyly and greater discontinuities between interspecific divergence and intraspecific variation (Hare, Citation2001; Palumbi et al., Citation2001; Zink & Barrowclough, Citation2008).