Figures & data
Fig. 1. Position of newly designed primers and generated overlapping amplicons for WRT329 (top), WRT345 (middle) and WRT349 (bottom) used to reconstruct up to 931 bp of tufA for C. floridana (Red: Thymine, Blue: Cytosine, Green: Adenine, Purple: Guanine). Note that amplicon tu157F-tu391R could not be amplified for WRT349 and is thus missing. Primer names are based on their position on the Ostreococcus tauri Courtes & Crétiennot complete tufA sequence (CR954199). Open boxes correspond to the multiple sequence alignments detailed in –.
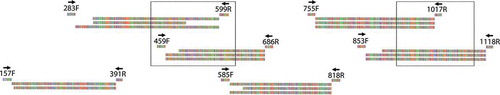
Table 1. Newly designed primers for the present study with position based on the Ostreococcus tauri Courtes & Crétiennot complete tufA sequence (CR954199). GC: Guanine-Cytosine content; Tm: melting temperature; bp: primer length in base pairs.
Table 2. Herbarium specimens of C. floridana sensu stricto present in MICH and examined in the present study. All originally collected from the Dry Tortugas, White Shoal, and off Southwest Channel.
Figs 2–3. Holotype and isotype of Caulerpa floridana W.R. Taylor. . Holotype of WRT361 maintained at MICH. Note early identification as C. ashmeadii Harvey. . Isotype of WRT361 maintained at NY. Note early identification as C. ashmeadii Harvey, later corrected as C. taylori M.A. Howe. Scale bars: 1 cm (–).
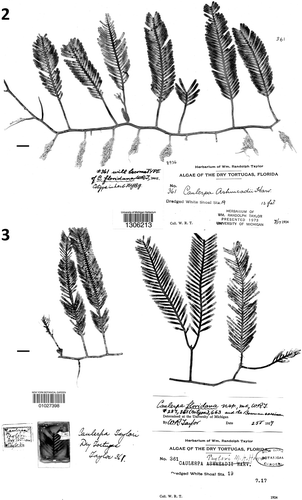
Figs 4–6. Historical herbarium specimens of Caulerpa floridana W.R. Taylor sampled for molecular assessment (note early identification as C. ashmeadii). . Specimen WRT349. . Specimen WRT329. . Specimen WRT345. All are maintained at MICH. Scale bars: 1 cm (–).
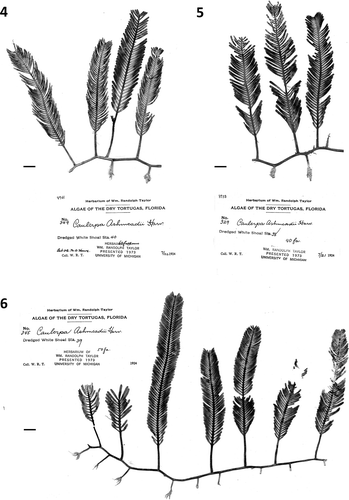
Fig. 7. Herbarium sheet of Caulerpa ashmeadii Harvey (TS1824) newly collected from the Florida Keys (Long Key) for DNA sequencing and morphological illustration. Scale bar: 1 cm.
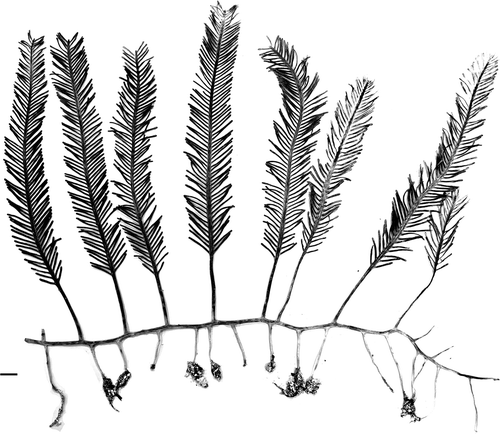
Fig. 8. Phylogenetic position of Caulerpa floridana W.R. Taylor and closely related species based on reconstructed tufA sequences for the three historical herbarium specimens studied, viz. WRT329 (931 bp), WRT345 (927 bp) and WRT349 (771 bp). The topology and bootstrap support values (numbers above branches) were obtained with RAxML. Labels in grey shade correspond to newly generated sequences. Lineages A–F represent entities of the C. racemosa-peltata complex sensu Sauvage et al. (Citation2013). Asterisks indicate alien strains of C. taxifolia.
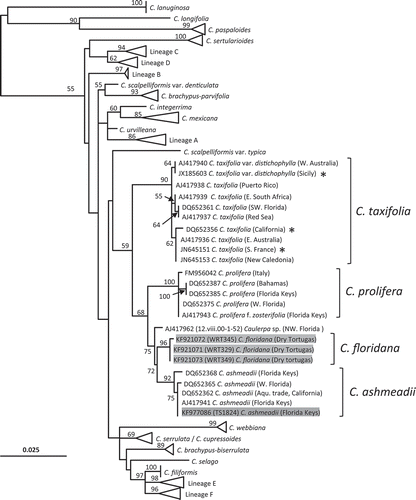
Fig. 9. Herbarium sheets of specimen SPF04257 reported as Caulerpa floridana W.R. Taylor by Joly et al. (Citation1976) from Ceará State, Brazil. Note pencil markings ‘floridana ?’ and ‘ashmeadii ?’ on the left and right thalli, respectively. Specimen maintained at SPF. Scale bar: 1 cm.
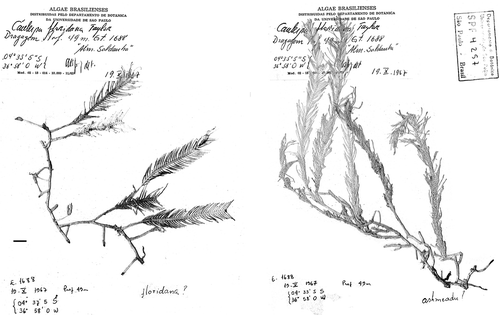
Table 3. Southwestern Atlantic specimens from Brazil reported as C. floridana or C. ashmeadii by Joly et al. (Citation1976) and Taylor (Citation1930).
Fig. 10. Herbarium sheets of specimen T13894 reported as Caulerpa ashmeadii Harvey by Taylor (Citation1930) from Pernambuco State, Brazil. See annotation made by Taylor regarding ‘traces of apiculi’ and pointed arrow on right assimilator. Specimen maintained at MICH Scale bar: 1 cm.

Fig. 11. Assimilator and pinnules comparison for C. floridana W.R. Taylor (WRT329, top) and C. ashmeadii Harvey (TS1824, bottom). Abbreviations: ap: apiculi; co: constrictions. Scale bar: 0.5 cm.
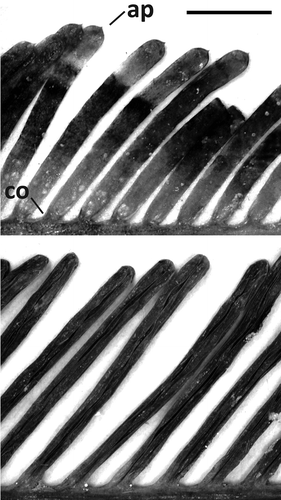
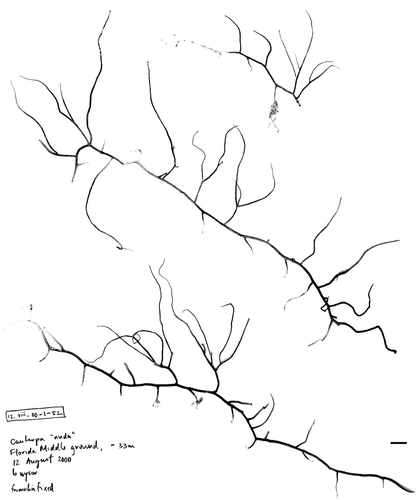
Fig. 12. Herbarium sheet of the unknown Caulerpa sp. (12.viii.00-1-52, AJ417962) originating from the Florida Middle Grounds previously sequenced by Famà et al. (2003). Specimen maintained at LAF. Scale bar: 1 cm.
Figs 13–14. Multiple sequence alignment of tufA detailing overlap between short amplicons generated for C. floridana specimens WRT329, WRT345 and WRT349, and sequence variation with closely related species C. ashmeadii, C. prolifera and C. sp. from the Florida Middle Ground (FMG). Conserved sites are shown as (.) in reference to C. prolifera. . Overlap between short amplicons tu283F-tu599R/tu459F-tu686R (position 244 to 394 of the analysed alignment in RAxML). Note site variation (Adenine: A) at position 355 (shaded in gray) for both amplicons generated for WRT345. Note low (lack) of variation on this stretch of tufA with sister species C. ashmeadii and C. sp. . Overlap between short amplicons tu755F-tu1017R/tu853F-tu1118R (position 719 to 870). Note nucleotide variation for C. floridana’s specimens at site 764 (Guanine: G) and 858 (Adenine: A) (shaded in grey).

