Figures & data
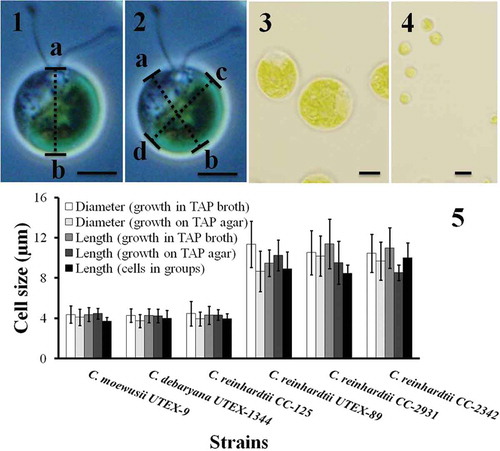
Figs 1–5. Cell size of Chlamydomonas strains grown under different conditions. Cell size was measured after cells were harvested from TAP agar, TAP broth and after they had formed groups. The cell size was measured in two different ways. In the first case the length from papilla to the posterior end (length ‘ab‘ in ) was measured. In the second case the cell diameter was measured at two different angles (the mean of diameter ‘ab‘ and ‘cd‘ in ). The results are mean ± SD; n = 40–50 cells of each strain. Chlamydomonas strains/species had different cell size (–): C. moewusii UTEX-9, C. debaryana UTEX-1344 and C. reinhardtii CC-125 were significantly smaller than C. reinhardtii UTEX-89, C. reinhardtii CC-2931 and C. reinhardtii CC-2342. A typical big cell and a small cell is shown in and respectively. Scale bars: –, 10 µm.
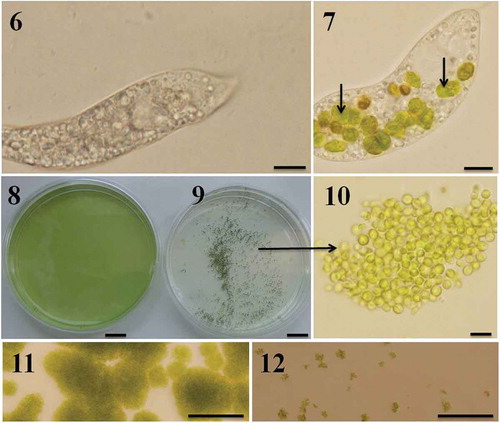
Figs 6–12. Predator-induced group formation in Chlamydomonas. Peranema raised in the laboratory using a mixture of spring water, boiled egg yellow and wheat grains is transparent and accumulates a large amount of starch granules (). Immediately after the Peranema cells were transferred into the suspension containing Chlamydomonas cells (in SVM medium) they started preying on green algal cells; intact Chlamydomonas cells are indicated with arrows (). Chlamydomonas cells formed groups soon after they were exposed to the predator. Shown here are C. moewusii UTEX-9 cells growing in the absence () and presence () of the predator, P. trichophorum. Note the groups formed by C. moewusii UTEX-9 cells in the presence of the predator (–). Predation was also studied on solid medium; Chlamydomonas cells were inoculated on SVM agar plates in the absence () and presence () of P. trichophorum and the plates were observed after 5 days of incubation at 22ºC. Since the motility of Chlamydomonas cells (not of Peranema) was restricted on solid plates they did not form groups and were heavily predated by Peranema (). In a control where Peranema was not added, algal growth was unaffected (). Scale bars: –, µm; –, cm; , 10 µm; –, 200 µm.
Figs 13–19. Group formation by Chlamydomonas in the presence of Peranema culture filtrate. Chlamydomonas cells were suspended in SVM and increasing amounts of Peranema culture filtrate added (0.5–4 ml; –). In a ‘control’ well () culture filtrate was not added; instead, the same amount of sterile, Peranema- naïve growth medium was added. In a ‘Peranema’ well () living Peranema cells were added. Chlamydomonas formed groups in the presence of Peranema culture filtrate and as the amount of filtrate was increased the number of cells in the groups (compared with free swimming) increased (). Shown here is a typical outcome from one of three such experiments in C. moewusii UTEX-9. In the mean values from three technical replicates from one experiment are plotted; coefficient of variation within an experiment is 5–15%. Scale bar: 1 cm.
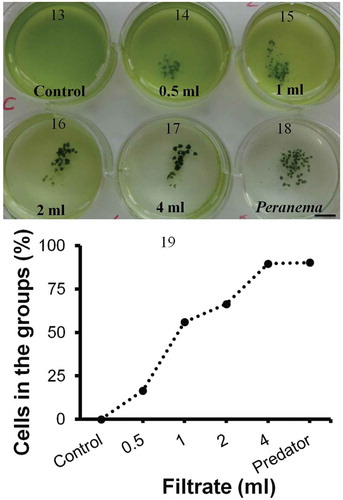
Table 1. Genetic similarity in Chlamydomonas reinhardtii strains. Genetic similarity was calculated from the published data (Kang & Fawley, Citation1997). The values in the table indicate the genetic similarity (%); in parentheses are the number of similar characters (DNA bands)/total characters (DNA bands).
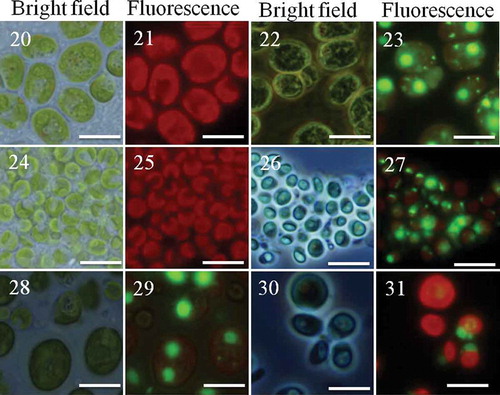
Figs 20–31. Clonal and chimaeric groups formed by Chlamydomonas in the presence of Peranema. Chlamydomonas cell cultures either as clones or binary mixes of two strains/species were made and inoculated with the predator Peranema. Chlamydomonas cells belonging to two different strains were identified based on their size and SYBR green I stain. Groups were formed after 18–48 h; the groups were observed under bright field or fluorescence (Green fluorescence protein filter). Shown here are groups formed by C. reinhardtii CC-2931(large cells) and C. moewusii UTEX-9 (small cells): – (C. reinhardtii CC-2931, unstained); – (C. reinhardtii CC-2931, stained); – (C. moewusii UTEX-9, unstained); – (C. moewusii UTEX-9, stained); – (chimaeric groups formed by a mix of C. reinhardtii CC-2931(large and stained cells) and small, unstained cells of C. moewusii UTEX-9); – (the chimaeric groups formed by the same pair except the stained strain was reversed; that is, small cells of C. moewusii UTEX-9 were stained and mixed with large, unstained cells of C. reinhardtii CC-2931). Scale bars: –, 10 µm.
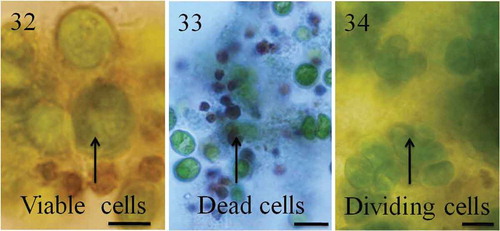
Figs 32–34. Cell turnover in groups. Chlamydomonas cells formed tight groups in the presence of predators or their culture filtrates. Within groups, some cells grew or divided, others died. Cells in the groups are protected from predators but for unknown reasons a large number of cells in the groups die. indicates green viable cells and dark brown dead cells. Cell death was confirmed using Trypan blue stain; dead cells appear blue as they fail to exclude Trypan blue (). Recently divided cells are shown in . Scale bars: –, 10 µm.