Figures & data
Table 1: Summary of the currently known introduced Pyropia species.
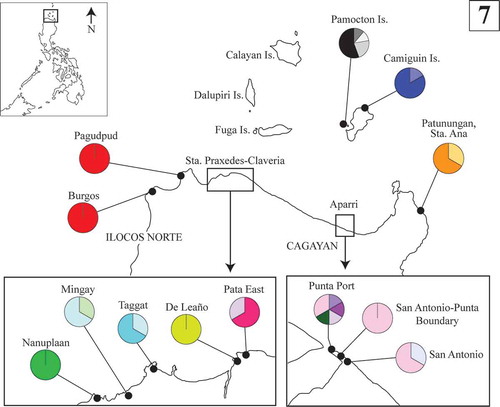
Table 2. Summary of sampling locations and haplotype information of Philippine Py. acanthophora specimens used in this study.
Table 3. Genetic diversity statistics of Py. acanthophora for each geographic location calculated for mitochondrial COI-5P and plastid rbcL sequences.