Figures & data
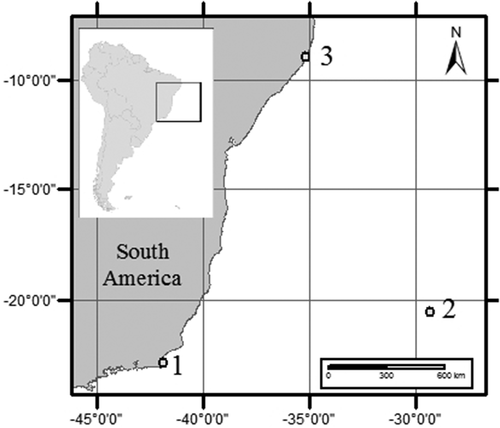
Fig. 1. Map of South America showing the three locations where species of Coolia were isolated from. 1 – Armação dos Búzios, Rio de Janeiro, 2 – Trindade Island, Espírito Santo, 3 – Maragogi, Alagoas, Brazil.
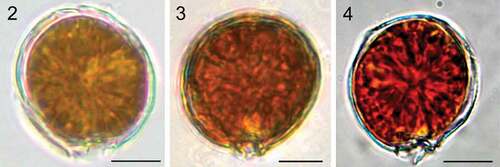
Figs 2–4. Light microscopy images. Fig. 2. Coolia malayensis (strain UNR-02). Fig. 3. Coolia tropicalis (strain UNR-28). Fig. 4. Coolia cf. canariensis phylogroup II (strain UNR-25). Scale bars: 10 µm.
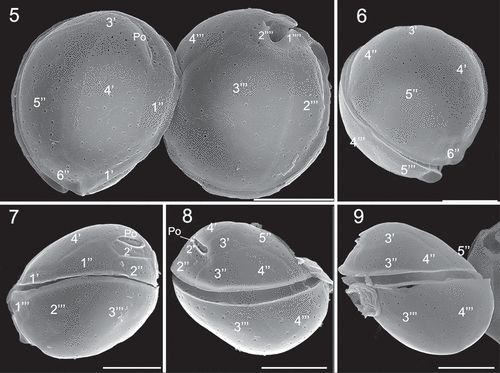
Figs 5–9. Scanning electron microscopy (SEM) micrographs of Coolia malayensis (strain UNR-02). Fig. 5. Apical and antapical view. Fig. 6. Oblique view of the epitheca. Fig. 7. Left lateral view. Fig. 8. Right lateral view. Fig. 9. Left-dorsal thecal plate view. Scale bars: 10 µm.
Figs 10–13. SEM micrographs of Coolia malayensis (strain UNR-02). Fig. 10. Detail of the apical pore complex showing the Po. Fig. 11. Detail of pores showing poroids. Fig. 12. Oblique view showing dorsal hypothecal plates. Fig. 13. Ventral view. Scale bars: (10–11) 2 µm, (12–13) 10 µm.
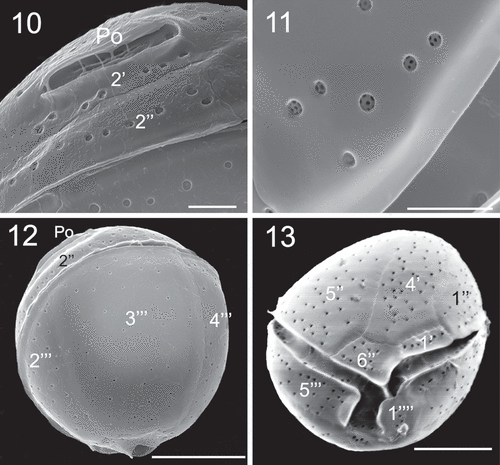
Table 1. Comparison of the morphological features of Coolia malayensis (strain UNR-02), C. tropicalis (strain UNR-28) and C. canariensis complex, including strain UNR-25
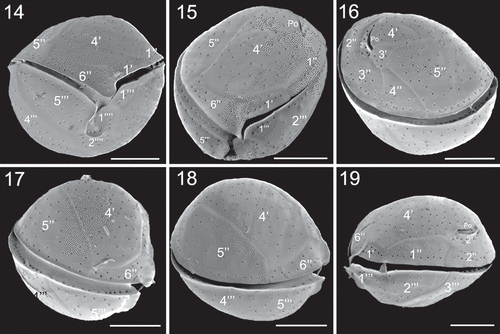
Figs 14–19. SEM micrographs of Coolia tropicalis (strain UNR-28). Fig. 14. Ventral view. Fig. 15. Oblique view of the epitheca. Fig. 16. Oblique-dorsal view of left side. Figs 17–18. Right lateral view. Fig. 19. Left lateral view. Scale bars: 10 µm.
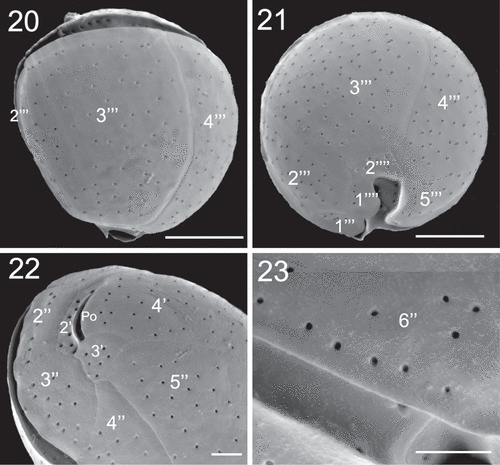
Figs 20–23. SEM micrographs of Coolia tropicalis (strain UNR-28). Fig. 20. Dorsal view of hypotheca. Fig. 21. Antapical view. Fig. 22. Left part of the epitheca in apical view. Fig. 23. Detail of pores on plate 6′′. Scale bars (20–21): 10 µm, (22–23): 3 µm.
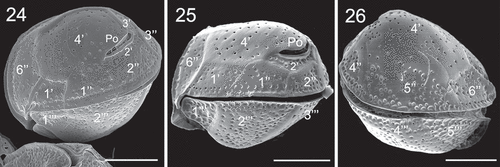
Figs 24–26. SEM micrographs of Coolia cf. canariensis phylogroup II. Fig. 24. Oblique-lateral view. Fig. 25. Left lateral view. Fig. 26. Right lateral view. Scale bars: 10 µm.
Figs 27–30. SEM micrographs of Coolia cf. canariensis phylogroup II. Fig. 27. Apical view. Fig. 28. Detail of the apical pore complex showing the Po. Figs 29–30. Inside view of the epitheca. Scale bars: (27, 29, 30): 10 µm; (28): 4 µm.
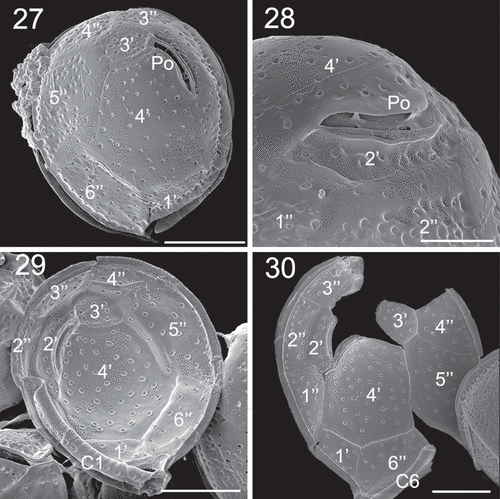
Figs 31–34. SEM micrographs of Coolia cf. canariensis phylogroup II. Figs 31–32. Oblique-dorsal view. Fig. 33. Antapical view. Fig. 34. Detail of the surface of plate 2′′′. Arrows: extensions of the thecal surface towards the cingulum. Scale bars: (31–33): 10 µm; (34): 4 µm.
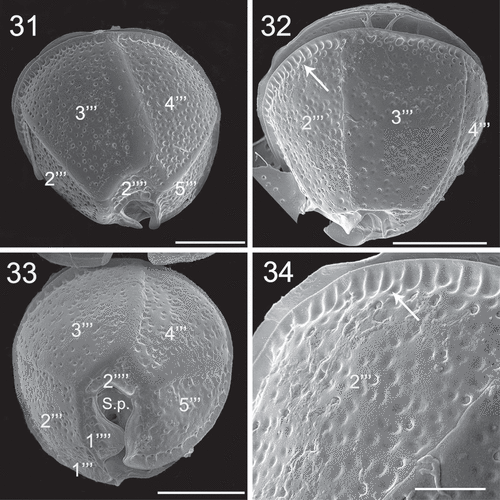
Figs 35–36. SEM micrographs of Coolia cf. canariensis phylogroup II. Fig. 35. Detail of plates surrounding the antapical part of the sulcal region. Fig. 36. Oblique-ventral view. Scale bars: 5 µm.
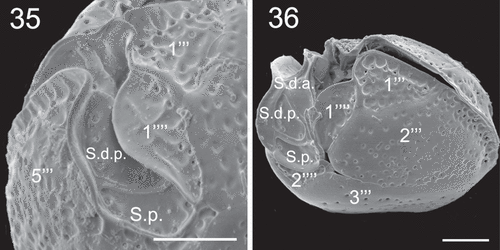
Figs 37–38. SEM micrographs of Coolia cf. canariensis phylogroup II in ventral view. Scale bars: 10 µm.
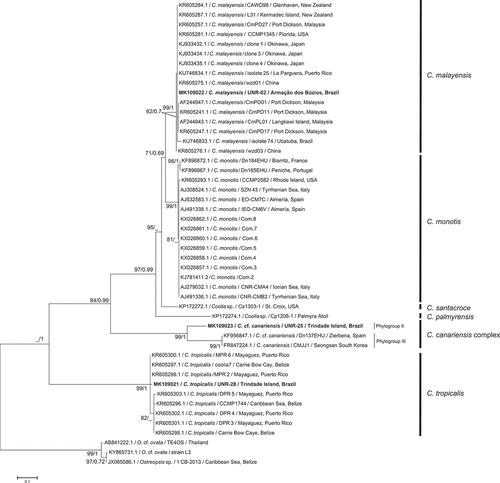
Fig. 39. Phylogenetic tree presenting consensus topology from Maximum likelihood analysis and Bayesian inference based on LSU sequences. Operational taxonomic units (OTUs) are identified by: GenBank acession number/species name/strain code/place of origin (when available). Numbers at nodes are bootstrap values from ML analysis and posterior probabilities from BI, respectively (cut-off = 50% for both analysis). New sequences published in this study are displayed in bold.
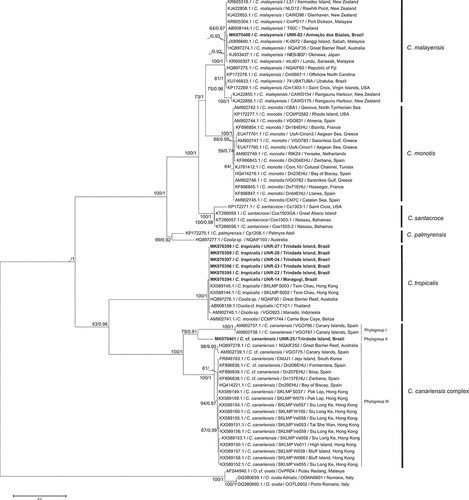
Fig. 40. Phylogenetic tree presenting consensus topology from Maximum likelihood analysis and Bayesian inference based on ITS sequences. Operational taxonomic units (OTUs) are identified by: GenBank acession number/species name/strain code/place of origin (when available). Numbers at nodes are bootstrap values from ML analysis and posterior probabilities from BI, respectively (cut-off = 50% for both analysis). New sequences published in this study are displayed in bold.