Figures & data
Fig. 1. Photograph of a culture box where Drosera capensis plants are grown in sterile conditions. Inset shows magnification of one of the plants, at the base of which a green biofilm can be observed (arrows).
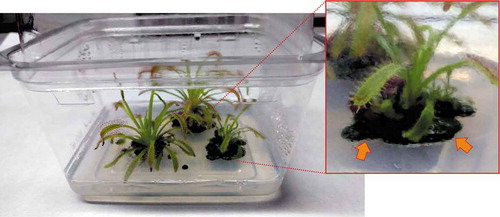
Fig. 2. Phylogenetic reconstruction of the genus Coccomyxa based on 18S rRNA gene analyses. The NJ topology is depicted and the numbers associated with nodes indicate support values for NJ, MP, ML and BI analyses, respectively. Only bootstrap supports ≥50% and posterior probabilities ≥0.95 are reported. Values for nodes that obtained support in only one of the performed phylogenetic analyses were omitted. Horizontal bar represents expected number of nucleotide substitutions per site. Grey boxes represent species according to the most recent classification or OTUs found/described in previous papers. Coccomyxa cimbrica sp. nov. is highlighted with a light blue box. Brackets plus white boxes report the classification by Darienko et al. (Citation2015).
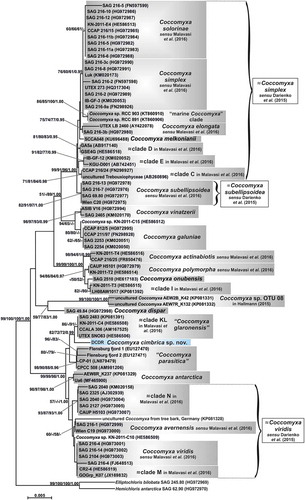
Fig. 3. Hypothetical secondary structure of the ITS2 spacer for Coccomyxa cimbrica sp. nov. The whole secondary structure is represented on a small scale in the centre of the figure; the nucleotide sequences of the spacers between the four main helices are reported on the structure. The 5.8S/LSU stem and the four main helices, indicated by dotted arrows, are depicted in detail at higher magnification; in paired regions, A-U pairings are represented with a single line, G-C pairings with a double line, and unconventional pairings with a line interrupted by a circle. Compensatory base changes (CBCs) between C. cimbrica and some focus taxa are represented by coloured squares (light blue for C. antarctica Ua6, red for strain UTEX SNO83, light green for strain KN-2011-C4) connected to the name of the interested taxon by double-headed black arrows.
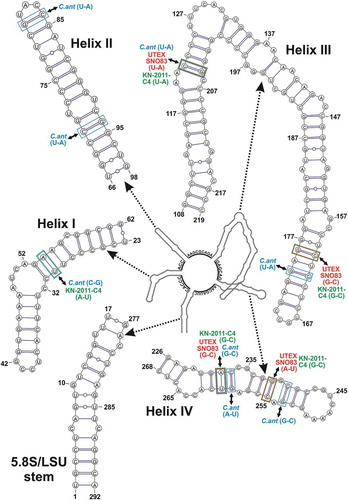
Fig. 4. Phylogenetic reconstruction of the genus Coccomyxa based on ITS2 spacer (sequences + structures) analyses. The NJ topology is depicted and the numbers associated with nodes indicate support values for NJ with ProfDistS, NJ with MEGA, MP and ML analyses, respectively. Only bootstrap supports ≥50% are reported. Values for nodes that obtained support in only one of the performed phylogenetic analyses were omitted. Horizontal bar represents expected number of nucleotide substitutions per site. Grey boxes represent species according to the most recent classification or OTUs found/described in previous papers. Coccomyxa cimbrica sp. nov. is highlighted with a black outline. Brackets plus white boxes report the classification by Darienko et al. (Citation2015).
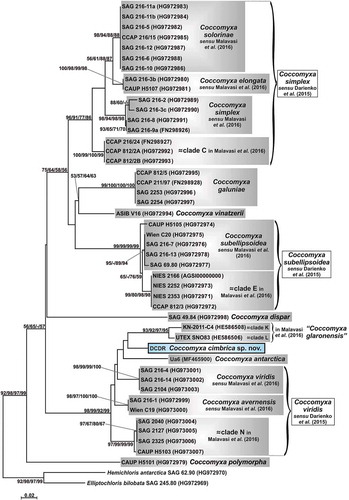
Fig. 5. Phylogenetic reconstruction of the genus Coccomyxa based on ITS2 numeric barcode analyses. The NJ topology is depicted and the numbers associated with nodes indicate support values for NJ, MP and ML analyses, respectively. Only bootstrap supports ≥50% are reported. Values for nodes that obtained support in only one of the performed phylogenetic analyses were omitted. Horizontal bar represents number of changes. Grey boxes represent species according to the most recent classification or OTUs found/described in previous papers. Coccomyxa cimbrica sp. nov. is highlighted with a black outline. Brackets plus white boxes show the classification by Darienko et al. (Citation2015). For each Coccomyxa strain, the corresponding ITS2 numeric barcode is reported.
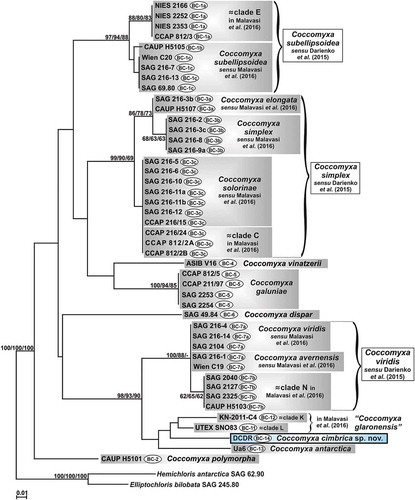
Figs 6–15. Morphology of Coccomyxa cimbrica sp. nov. Figs 6–10. Variable shapes of culture cells. Fig. 11. Cell reproduction by oblique division (arrow). Figs 12–13. Asexual reproduction through cell division with the formation of 2–4 autospores. Figs 14–15. Fluorescence images of culture cells stained with 4′,6 diamidino-2-phenylindole (DAPI). Note small mucilaginous caps (double arrows), reproduction by oblique division (arrow head) and, in Fig. 14, an autosporangium (arrow). Scale bars: Figs 6–13, 2 µm; Figs 14–15, 3 µm.
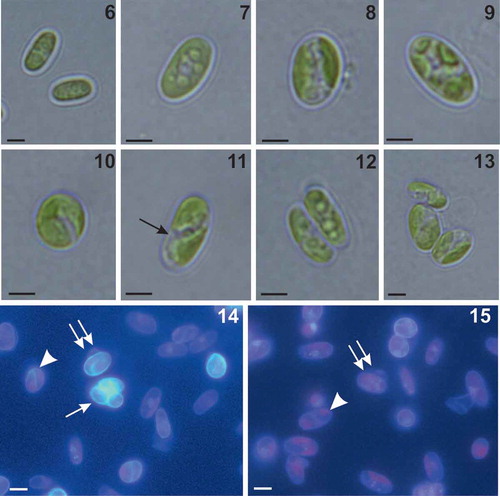
Figs 16–22. Morphology and ultrastructure of Coccomyxa cimbrica sp. nov. Fig. 16. Scanning electron micrograph of elliptical vegetative cells showing smooth cell wall. Figs 17, 18. Scanning electron micrographs of asexual reproduction by cell division through the rupture of mother cell wall and the formation of 2 (Fig. 17) and 3 (Fig. 18) autospores. Figs 19–20. Ultrastructure of cells showing thin and smooth cell wall (cw), nucleus (n), mitochondria (m), and a parietal chloroplast (ch) lacking pyrenoid, with some starch granules (s). Note in the cytoplasm the presence of some electron dense bodies (ei) and lipid bodies (lb). Fig. 21. Detail of the thylakoid membranes (t), single or piled up forming grana, and a starch granule (s) located among the thylakoids. Fig. 22. Micrograph of an autosporangium with four mature vegetative daughter cells enclosed by the mother cell wall (cw). Note in the autospores the presence of chloroplast (ch) and nucleus (n). Scale bars: Figs 16, 17, 18, 2 µm; Fig. 19, 1 µm; Fig. 20, 500 nm; Fig. 21, 200 nm; Fig. 22, 2 µm.
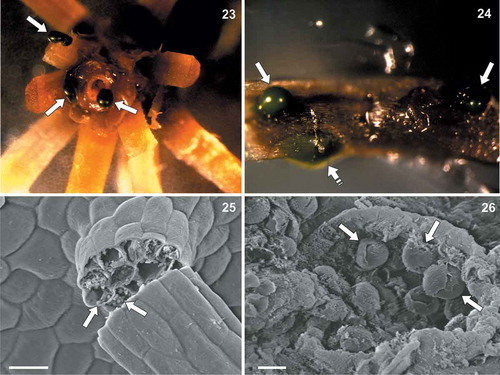
Figs 23–26. Coccomyxa cimbrica sp. nov. associated with Drosera capensis plants. Fig. 23. Stereomicroscope magnification of a sterilized D. capensis stem fragment after 70% ethanol + sodium hypochlorite washings. Green masses corresponding to C. cimbrica colonies are observed on the plant surface and coming out from the stem (white arrows). Fig. 24. Stereomicroscope magnification of a sterilized D. capensis leaf portion. Green C. cimbrica colonies are observed just below the plant epidermis (white dashed arrow) and coming out from tissue lacerations (white arrows). Fig. 25. Scanning electron micrograph of a D. capensis leaf sticky glandular hair (‘tentacle’). Inside the broken tentacle, globular structures resembling C. cimbrica cells are present (white arrows). Fig. 26. Scanning electron micrograph of D. capensis leaf surface. From a tissue laceration, globular structures similar to C. cimbrica cells are observed (white arrows). Scale bars: Fig. 16, 20 µm; Fig. 17, 5 µm.