Figures & data
Fig. 1. Hidden diversity and host range in the marine genus Olpidiopsis. Phylogenetic anchoring of all V4 18S rDNA Operational Taxonomic units (OTUs) identified in this study with reference Olpidiopsis sequences, as well as four additional environmental 18S sequences (KT815133, KT810401, AY426928, AY789783) identified by similarity search in GenBank. Grey brackets define the three Olpidiopsis lineages named after their first described species. Olpidiopsis species initially isolated on Bangiophycean hosts are indicated in red. Purple, green and blue OTUs correspond to sequences obtained from P. umbilicalis metabarcoding, the Australian Marine Dataset (AMD) and our dedicated SRA screening pipeline, respectively. The right panel summarizes the host range of Olpidiopsis spp. based on previous laboratory experiments, where hosts can be infected (+), not infected (−), not tested (NT), or probably infected based on metabarcoding evidence (*). Dashed circles highlight naturally occurring hosts.
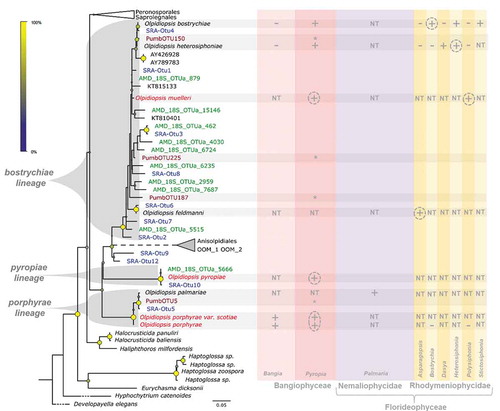
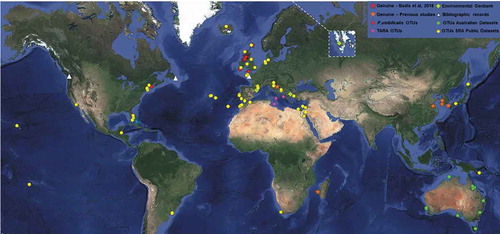
Fig. 2. Overview of biogeographic evidence in the marine genus Olpidiopsis. Worldwide evidence of the occurrence of marine Olpidiopsis parasites, including the GPS coordinates of all molecular data () collected in this study, additional morphological reports of marine Olpidiopsis and a selection of red algal pathogens of doubtful taxonomic position (inland points represent missing locations), summarized in Supplementary table S1. Stars: Genuine Olpidiopsis isolates; Squares: GenBank environmental sequences; Triangles: morphological reports; Dots: Operational Taxonomic Units gathered from the Sequence Read Archive (yellow), Tara Oceans (purple), the Australian Marine Dataset (green) and P. umbilicalis microbiome (red). Individual maps of each SRA-OTUs are provided in Supplementary figs S2–S13.
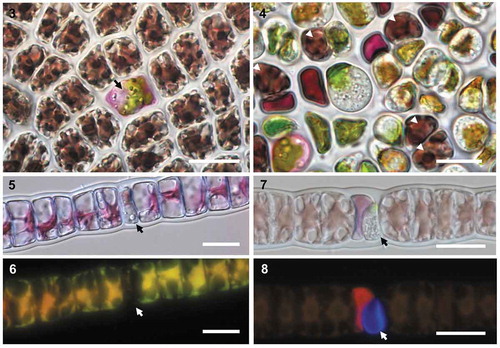
Figs 3–8. Non-native Olpidiopsis parasites could infect wild and farmed red algae. Cross-inoculations of Korean and Scottish algae with non-native Olpidiopsis parasites. Figs 3–4. O. porphyrae var. scotiae infecting the major Korean commercial cultivar of Pyropia yezoensis. Fig. 3. Early developing parasite thallus (arrow) 48 hours after infection. Fig. 4. Densely infected blade of the same Pyropia cultivar 5 days after infection, where only 5 host cells are left alive (white arrowheads). Figs 5–8. Young (5–6) and mature (7–8) thalli of the Korean parasite Olpidiopsis porphyrae var. koreanae (arrow) infecting Bangia sp. from Scotland. As revealed by Calcofluor White staining, the early stage of thallus development is unwalled (6) while a clear cell wall is detected in mature thallus (7). Scale bars: 20 μm.