Figures & data
Table 1. Characiopsis strains studied and their characteristics. Cell morphology refers to the cell forms that predominate in the strains studied, with the caveat that the morphology can vary in the same culture. Dimensions of the cell body and stipe correspond to length × width (the general range is indicated, with exceptional values in parentheses)
Figs 1–8. Characiopsis strains with morphology similar to Characiopsis minuta and unidentified Characiopsis strains. Fig. 1. Characium minutum (adapted from Braun, Citation1855). Fig. 2. Characiopsis minuta ACOI 2423. Fig. 3. Characiopsis cf. minuta ACOI 2425. Fig. 4. Characiopsis sp. ACOI 2432. Fig. 5. Characiopsis sp. ACOI 2438B. Fig. 6. Characiopsis sp. ACOI 2429. Fig. 7. Characiopsis sp. ACOI 2429A. Fig. 8. Characiopsis sp. ACOI 2430. Old cell (o c), pyrenoid (py), reddish globule (r g), stipe (st), zoospore (z), zoosporangium (zs). Photos 2–10 DIC 100× APO. Scale bars: 10 μm

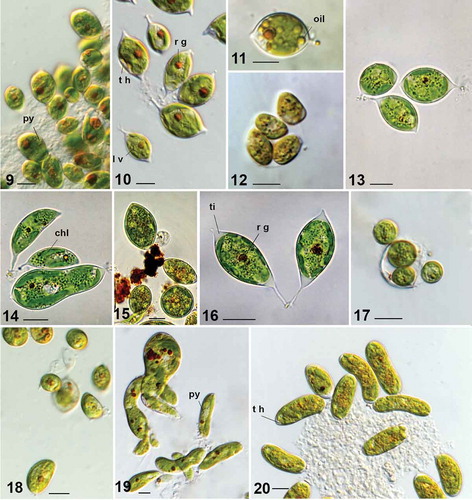
Figs 9–20. Characiopsis strains with morphology unlike that of Characiopsis minuta. Fig. 9. Characiopsis pernana ACOI 2433. Figs 10–12. Characiopsis acuta ACOI 456. Fig. 13. Characiopsis acuta ACOI 1837. Fig. 14. Characiopsis longipes ACOI 1838. Fig. 15. Characiopsis longipes ACOI 1838. Fig. 16. Characiopsis longipes ACOI 2438. Fig. 17. Characiopsis longipes ACOI 1839_9. Fig. 18. Characiopsis cf. minutissima ACOI 2427A. Fig. 19. Characiopsis cf. saccata ACOI 481. Fig. 20. Characiopsis cedercreutzii ACOI 3169. Apical tip (ti), chloroplast (chl), lamellate vesicles (l v), oil droplets (oil), pyrenoid (py), reddish globule (r g), cell wall thickening (t h). Photos 11–14 and 19–22 DIC 100× APO. Scale bars: 10 μm
Figs 21–24. TEM sections of Characiopsis vegetative cells. Fig. 21. C. minuta ACOI 2423. Figs 22–24. C. cf. saccata ACOI 481. Chloroplast (chl), Golgi body (G b), lamellate vesicles (l v), mitochondrion (m), nucleus (n), nucleolus (nu), pyrenoid (py), reddish globule (r g). Scale bars: 1 μm
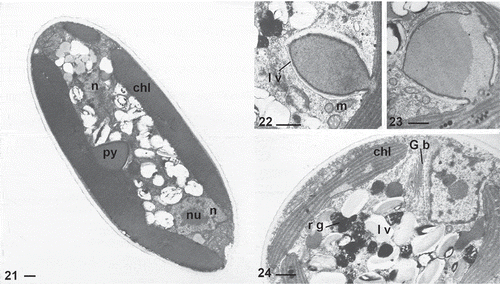
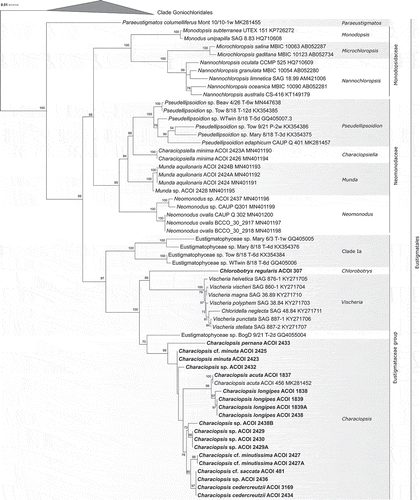
Fig. 25. Phylogeny of Eustigmatophyceae based on sequences of the 18S rRNA gene, showing the Eustigmatales. The phylogeny shown was inferred using the Maximum likelihood method implemented in RAxML (employing GTR+Γ substitution model) with bootstrap analysis followed by a thorough search for the ML tree. Bootstrap values higher than 50 are shown. Labels at terminal leaves comprise the strain updated taxonomic name followed by the collection reference number and the GenBank accession number. New sequences are highlighted in bold. The tree was rooted using 15 sequences from stramenopile algae sampled from GenBank. The outgroup is omitted and the ordinal clade Goniochloridales is shown collapsed for simplicity
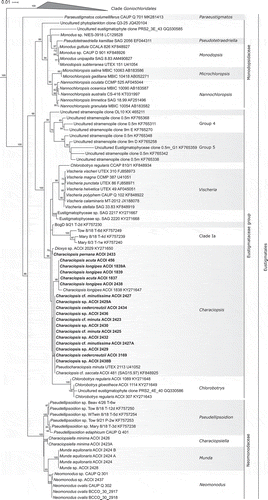
Fig. 26. Phylogeny of Eustigmatophyceae based on the rbcL gene, showing the Eustigmatales. The phylogeny shown was inferred using the Maximum likelihood method implemented in RAxML (employing GTR+Γ substitution model) with bootstrap analysis followed by a thorough search for the ML tree. Bootstrap values higher than 50 are shown. Labels at terminal leaves comprise the strain updated taxonomic name followed by the collection reference number when applicable and the GenBank accession number. New sequences highlighted in bold. The tree was rooted at the ordinal clade Goniochloridales which is shown collapsed for simplicity