Figures & data
Table 1. The composition of TAP media and nitrogen deficient media
Figure 1. Comparison of physiological and biochemical indicators of P_KND (Parachlorella kessleri nitrogen deficient), and P_KC (P. kessleri control) over 7 days of cultivation. (a) Dry weight (bar graph) and neutral lipid content (line). (b) chlorophyll content (bar graph) and photochemical efficiency of photosystem II (Fv/Fm) (line). Data are shown as mean ± SE, n = 3
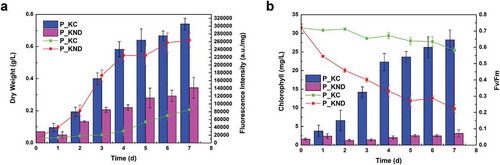
Table 2. Biochemical indicators of P_KND (nitrogen deficient) and P_KC (control) at day 0 and day 7 of cultivation
Table 3. The fatty acid compositions of P_KND and P_KC on day 7
Table 4. The output of transcriptome sequencing of Parachlorella kessleri TY
Table 5. Splicing length distribution of unigenes in Parachlorella kessleri TY