Figures & data
Figure 1. Effects of PI3K or PKC inhibition on HDL-lipid uptake mediated by human or mouse SR-BI. LdlA[hSR-BI] cells (Panels A and C) and ldlA[mSR-BI] cells (Panels B and D), were treated without (black bars) or with (grey bars) increasing concentrations of either wortmannin (Panels A and B) or Ro31-8220 (Panels C and D) for 10 min prior to and during incubation for 2 h at 37°C with 5 µg/ml DiI-HDL. Untransfected ldlA7 cells (white bars) were incubated with DiI-HDL in parallel. Cells were washed and fluorescence in each well was determined using a cytofluor 3000 fluorescence plate reader as described in the Methods section. DiI-uptake is expressed as relative fluorescence units normalized to the amount of protein in each well. Results are averages±standard deviations of three determinations.
![Figure 1. Effects of PI3K or PKC inhibition on HDL-lipid uptake mediated by human or mouse SR-BI. LdlA[hSR-BI] cells (Panels A and C) and ldlA[mSR-BI] cells (Panels B and D), were treated without (black bars) or with (grey bars) increasing concentrations of either wortmannin (Panels A and B) or Ro31-8220 (Panels C and D) for 10 min prior to and during incubation for 2 h at 37°C with 5 µg/ml DiI-HDL. Untransfected ldlA7 cells (white bars) were incubated with DiI-HDL in parallel. Cells were washed and fluorescence in each well was determined using a cytofluor 3000 fluorescence plate reader as described in the Methods section. DiI-uptake is expressed as relative fluorescence units normalized to the amount of protein in each well. Results are averages±standard deviations of three determinations.](/cms/asset/409d47a0-a81c-42a5-9ded-0310a269d908/imbc_a_229945_f0001_b.gif)
Figure 2. Effects of inhibition of endocytosis on HDL-lipid uptake by human or mouse SR-BI. LdlA[hSR-BI] or ldlA[mSR-BI] were either depleted of intracellular potassium, exposed to hypertonic sucrose, 10 µM cytochalasin D or 5 mM NaN3 and 50 mM 2-deoxyglucose (Energy Depletion) as described in the Methods section. A-E: Alexa 594 transferrin uptake (red fluorescence) by ldlA[hSR-BI] cells was monitored as a control for endocytosis. Cell nuclei were stained with DAPI (blue fluorescence). Representative images are shown. Equivalent results were obtained for ldlA[mSR-BI] cells (not shown). Scale bars = 10 µm. Panels F–I: Cells were either depleted of potassium (panels F and H) or exposed to hypertonic sucrose (panels G and I) prior to and during incubation with 5 µg/ml of DiI-HDL for 2 h at 37°C in the absence or presence of 200 µg/ml of maleyl-BSA (a general competitor for SR-BI-mediated lipoprotein lipid uptake). Cellular uptake of DiI was measured by flow cytometry. Specific uptake of DiI was determined as the difference in the mean fluorescence values of the population of cells incubated without and with maleyl-BSA (equivalent results were obtained when unlabeled lipoproteins were used instead of maleyl-BSA). Values were normalized to control, untreated ldlA[hSR-BI] (panels F and G; black bars, set at 100% for ease of comparison) or ldlA[mSR-BI] cells (panels H and I; black bars, set at 100% for ease of comparison). Grey bars correspond to treated cells and white bars correspond to untreated control untrasnfected ldlA7 cells. Data are averages±standard deviations of triplicates. Stars (*) indicate p<0.05 for comparisons with untreated ldlA[hSR-BI] or ldlA[mSR-BI] cells by Student's t-test (Microsoft Excel).
![Figure 2. Effects of inhibition of endocytosis on HDL-lipid uptake by human or mouse SR-BI. LdlA[hSR-BI] or ldlA[mSR-BI] were either depleted of intracellular potassium, exposed to hypertonic sucrose, 10 µM cytochalasin D or 5 mM NaN3 and 50 mM 2-deoxyglucose (Energy Depletion) as described in the Methods section. A-E: Alexa 594 transferrin uptake (red fluorescence) by ldlA[hSR-BI] cells was monitored as a control for endocytosis. Cell nuclei were stained with DAPI (blue fluorescence). Representative images are shown. Equivalent results were obtained for ldlA[mSR-BI] cells (not shown). Scale bars = 10 µm. Panels F–I: Cells were either depleted of potassium (panels F and H) or exposed to hypertonic sucrose (panels G and I) prior to and during incubation with 5 µg/ml of DiI-HDL for 2 h at 37°C in the absence or presence of 200 µg/ml of maleyl-BSA (a general competitor for SR-BI-mediated lipoprotein lipid uptake). Cellular uptake of DiI was measured by flow cytometry. Specific uptake of DiI was determined as the difference in the mean fluorescence values of the population of cells incubated without and with maleyl-BSA (equivalent results were obtained when unlabeled lipoproteins were used instead of maleyl-BSA). Values were normalized to control, untreated ldlA[hSR-BI] (panels F and G; black bars, set at 100% for ease of comparison) or ldlA[mSR-BI] cells (panels H and I; black bars, set at 100% for ease of comparison). Grey bars correspond to treated cells and white bars correspond to untreated control untrasnfected ldlA7 cells. Data are averages±standard deviations of triplicates. Stars (*) indicate p<0.05 for comparisons with untreated ldlA[hSR-BI] or ldlA[mSR-BI] cells by Student's t-test (Microsoft Excel).](/cms/asset/8475c019-7968-4bdc-bfc8-de021ccce369/imbc_a_229945_f0002_b.jpg)
Figure 3. Effects of energy depletion on human or mouse SR-BI cell surface localization and HDL binding. LdlA[hSR-BI] and ldlA[mSR-BI] cells were subjected to energy depletion or control treatment as described in the Methods section. Cells were chilled to 4°C, and either released from dishes for cell surface biotinylation (panel A) or incubated with 5 µg/ml DiI-HDL for 1 h at 4°C to monitor HDL binding (panel B). (A) Cell surface biotinylation, protein solubilization and streptavidin pull-down were performed as described in the Methods section. One half of the pull-down sample (top panel) and total cell lysate equivalent to 1/70 of the pull down sample (bottom panel) were analyzed by SDS-PAGE (10% acrylamide) and immunoblotting with an antibody that recognizes the C-terminus of SR-BI and a donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase. Detection was by enhanced chemiluminescence. (B) Control (black bars) or energy depleted (grey bars) ldlA[hSR-BI] or ldlA[mSR-BI] or untreated control ldlA7 cells (white bars) were incubated with 5 µg/ml DiI-HDL for 1 h at 4°C, prior to washing and analysis of DiI-fluorescence using a fluorescence plate reader. Values are the means±standard deviations of triplicate samples and are expressed as the percent binding relative to untreated ldlA[hSR-BI] or ldlA[mSR-BI] cells.
![Figure 3. Effects of energy depletion on human or mouse SR-BI cell surface localization and HDL binding. LdlA[hSR-BI] and ldlA[mSR-BI] cells were subjected to energy depletion or control treatment as described in the Methods section. Cells were chilled to 4°C, and either released from dishes for cell surface biotinylation (panel A) or incubated with 5 µg/ml DiI-HDL for 1 h at 4°C to monitor HDL binding (panel B). (A) Cell surface biotinylation, protein solubilization and streptavidin pull-down were performed as described in the Methods section. One half of the pull-down sample (top panel) and total cell lysate equivalent to 1/70 of the pull down sample (bottom panel) were analyzed by SDS-PAGE (10% acrylamide) and immunoblotting with an antibody that recognizes the C-terminus of SR-BI and a donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase. Detection was by enhanced chemiluminescence. (B) Control (black bars) or energy depleted (grey bars) ldlA[hSR-BI] or ldlA[mSR-BI] or untreated control ldlA7 cells (white bars) were incubated with 5 µg/ml DiI-HDL for 1 h at 4°C, prior to washing and analysis of DiI-fluorescence using a fluorescence plate reader. Values are the means±standard deviations of triplicate samples and are expressed as the percent binding relative to untreated ldlA[hSR-BI] or ldlA[mSR-BI] cells.](/cms/asset/f471a1d2-f898-427c-984f-3cf924db0a29/imbc_a_229945_f0003_b.gif)
Figure 4. Inhibition of endocytosis increases the efficiency of selective HDL-lipid uptake mediated by hSR-BI. LdlA[hSR-BI] cells were either not treated (‘Control’ and ‘ + HDL’) or subjected to energy depletion (ED), potassium depletion, or treatment with cytochalasin D or 1 µM wortmannin (as indicated). DiI/alexa 488-double labeled HDL (5 µg/ml) was added without or with 200 µg/ml unlabeled HDL (‘ + HDL’). After a 2 h incubation at 37°C, two-color flow cytometric analysis was used to measure uptake of both DiI (Panel A – ‘Lipid Uptake’) and alexa 488 (Panel B – ‘Protein Uptake’). (C) For each sample, the ratio of DiI /alexa 488 fluorescence of the cell population was determined as a measure of the ratio of HDL lipid/protein uptake. Data are the averages±standard deviations of three independent samples.
![Figure 4. Inhibition of endocytosis increases the efficiency of selective HDL-lipid uptake mediated by hSR-BI. LdlA[hSR-BI] cells were either not treated (‘Control’ and ‘ + HDL’) or subjected to energy depletion (ED), potassium depletion, or treatment with cytochalasin D or 1 µM wortmannin (as indicated). DiI/alexa 488-double labeled HDL (5 µg/ml) was added without or with 200 µg/ml unlabeled HDL (‘ + HDL’). After a 2 h incubation at 37°C, two-color flow cytometric analysis was used to measure uptake of both DiI (Panel A – ‘Lipid Uptake’) and alexa 488 (Panel B – ‘Protein Uptake’). (C) For each sample, the ratio of DiI /alexa 488 fluorescence of the cell population was determined as a measure of the ratio of HDL lipid/protein uptake. Data are the averages±standard deviations of three independent samples.](/cms/asset/fe89e238-d3eb-4ded-b09d-e24793cdf195/imbc_a_229945_f0004_b.gif)
Figure 5. Expression and activity of eGFP-tagged human SR-BI in transfected ldlA7 cells. (A) LdlA[mSR-BI] (lane 1) and ldlA7 cells (lanes 2, 3 and 5) were cultured in medium containing 5% FBS. LdlA7 cells were either not transfected (lane 2) or transfected with hSR-BI (lane 3) or eGFP-hSR-BI expression plasmids (lane 5). After 3 days, cell lysates were prepared and analyzed by SDS-PAGE and immunoblotting for SR-BI. Lane 4 is empty. (B) Cells transfected with the eGFP-hSR-BI expression plasmid were subjected to G418 selection and cell sorting as described in Materials and Methods, to generate ldlA[eGFP-hSR-BI] cells that stably overexpress eGFP-hSR-BI. Untransfected ldlA7 (grey histogram) and ldlA[eGFP-hSR-BI] (black histogram) cells were incubated with 5 µg/ml DiI-HDL for 2 h at 37°C. DiI uptake was measured by flow cytometry as described in the Methods section. Representative histograms are shown.
![Figure 5. Expression and activity of eGFP-tagged human SR-BI in transfected ldlA7 cells. (A) LdlA[mSR-BI] (lane 1) and ldlA7 cells (lanes 2, 3 and 5) were cultured in medium containing 5% FBS. LdlA7 cells were either not transfected (lane 2) or transfected with hSR-BI (lane 3) or eGFP-hSR-BI expression plasmids (lane 5). After 3 days, cell lysates were prepared and analyzed by SDS-PAGE and immunoblotting for SR-BI. Lane 4 is empty. (B) Cells transfected with the eGFP-hSR-BI expression plasmid were subjected to G418 selection and cell sorting as described in Materials and Methods, to generate ldlA[eGFP-hSR-BI] cells that stably overexpress eGFP-hSR-BI. Untransfected ldlA7 (grey histogram) and ldlA[eGFP-hSR-BI] (black histogram) cells were incubated with 5 µg/ml DiI-HDL for 2 h at 37°C. DiI uptake was measured by flow cytometry as described in the Methods section. Representative histograms are shown.](/cms/asset/4a4a6614-4e0a-4e63-9b26-74c26c1ac562/imbc_a_229945_f0005_b.gif)
Figure 6. Localization and dynamics of eGFP- hSR-BI. LdlA[eGFP-hSR-BI], ldlA[mRFP-hSR-BI] and ldlA[mSR-BI] cells were cultured on poly-D-lysine-coated glass coverslips fixed to 35 mm culture dishes. Cells were fixed with 2.5% paraformaldehyde and either visualized directly (panels A and B) or after permeabilization and indirect immunofluorescence using anti-SR-BI495 antibody and alexa 488-anti-rabbit secondary antibody (panel C) using wide-field fluorescence microscopy with standard rhodamine and FITC filter sets. Panels D–F correspond to DIC images of the cells shown in A–C. Scale bars = 10 µm. (G–J) Live-cell confocal imaging of ldlA[eGFP-hSR-BI] cells was performed as described in the Methods section. Panel G on the left shows three adjacent ldlA[eGFP-hSR-BI] cells. Scale bar = 10 µm. The white boxes represent regions that are magnified in panels H–J. Panel H shows a time series at 50 sec intervals, demonstrating eGFP-hSR-BI dynamics associated with a tether between two adjacent cells. The first panel (time 0 sec) is cropped and magnified from panel G. The arrow serves as a reference marker to illustrate the leftward motion of eGFP-hSR-BI fluorescence along the tether. Scale bar = 5 µm. This series of images corresponds to Supplementary Video 1 – online version only. Panel I shows images at 10 sec intervals, beginning at time 115 sec relative to the image in panel G. They illustrate the motion of an eGFP-hSR-BI-positive puncta (small box) towards the center of the cell. The arrow indicates an eGFP-hSR-BI-positive structure that is static within the time-frame of the images. Scale bar = 5 µm. This series of images corresponds to Supplementary Video 2 – online version only. Panel J shows a series of images taken at 5 sec intervals and focusing on the region of cell-cell contact between two adjacent cells. The cell on the upper left in this panel corresponds to the cell in the lower right in panels H and I (see panel G). Scale bar = 2.5 µm. The arrow indicates the position at which an SR-BI positive puncta, apparently derived from the cell surface, appears. This corresponds to Supplementary Video 3 – online version only.
![Figure 6. Localization and dynamics of eGFP- hSR-BI. LdlA[eGFP-hSR-BI], ldlA[mRFP-hSR-BI] and ldlA[mSR-BI] cells were cultured on poly-D-lysine-coated glass coverslips fixed to 35 mm culture dishes. Cells were fixed with 2.5% paraformaldehyde and either visualized directly (panels A and B) or after permeabilization and indirect immunofluorescence using anti-SR-BI495 antibody and alexa 488-anti-rabbit secondary antibody (panel C) using wide-field fluorescence microscopy with standard rhodamine and FITC filter sets. Panels D–F correspond to DIC images of the cells shown in A–C. Scale bars = 10 µm. (G–J) Live-cell confocal imaging of ldlA[eGFP-hSR-BI] cells was performed as described in the Methods section. Panel G on the left shows three adjacent ldlA[eGFP-hSR-BI] cells. Scale bar = 10 µm. The white boxes represent regions that are magnified in panels H–J. Panel H shows a time series at 50 sec intervals, demonstrating eGFP-hSR-BI dynamics associated with a tether between two adjacent cells. The first panel (time 0 sec) is cropped and magnified from panel G. The arrow serves as a reference marker to illustrate the leftward motion of eGFP-hSR-BI fluorescence along the tether. Scale bar = 5 µm. This series of images corresponds to Supplementary Video 1 – online version only. Panel I shows images at 10 sec intervals, beginning at time 115 sec relative to the image in panel G. They illustrate the motion of an eGFP-hSR-BI-positive puncta (small box) towards the center of the cell. The arrow indicates an eGFP-hSR-BI-positive structure that is static within the time-frame of the images. Scale bar = 5 µm. This series of images corresponds to Supplementary Video 2 – online version only. Panel J shows a series of images taken at 5 sec intervals and focusing on the region of cell-cell contact between two adjacent cells. The cell on the upper left in this panel corresponds to the cell in the lower right in panels H and I (see panel G). Scale bar = 2.5 µm. The arrow indicates the position at which an SR-BI positive puncta, apparently derived from the cell surface, appears. This corresponds to Supplementary Video 3 – online version only.](/cms/asset/d2eb3838-a105-498a-907a-085bab518a8e/imbc_a_229945_f0006_b.gif)
Figure 7. mRFP-tagged hSR-BI does not co-localize with lysosomal markers or with clathrin. LdlA[mRFP-hSR-BI] cells (panels A–F) and ldlA7 cells stably expressing both eGFP-clathrin and mRF-hSR-BI (panels G–I) were cultured on glass cover slips and fixed. Cells were permeabilized and immunostained for lysosomal markers cathepsin A (panel A) or saposin A (panel D). Alexa 488 labeled secondary antibody was used for detection prior to wide-field epifluorescence microscopy using a Zeiss Axiovert 200 M inverted microscope. Alternatively, cells were imaged directly (panels G–I) using a Zeiss LSM510 laser scanning confocal microscope. A, D and G: Alexa 488 or eGFP fluorescence. B, E and H: mRFP fluorescence. C, F and I: merged color images in which green corresponds to cathepsin A (panel C), saposin A (panel F) or eGFP-clathrin (panel I) and red corresponds to mRFP-hSR-BI. Scale bars = 10 µm. Representative images are shown.
![Figure 7. mRFP-tagged hSR-BI does not co-localize with lysosomal markers or with clathrin. LdlA[mRFP-hSR-BI] cells (panels A–F) and ldlA7 cells stably expressing both eGFP-clathrin and mRF-hSR-BI (panels G–I) were cultured on glass cover slips and fixed. Cells were permeabilized and immunostained for lysosomal markers cathepsin A (panel A) or saposin A (panel D). Alexa 488 labeled secondary antibody was used for detection prior to wide-field epifluorescence microscopy using a Zeiss Axiovert 200 M inverted microscope. Alternatively, cells were imaged directly (panels G–I) using a Zeiss LSM510 laser scanning confocal microscope. A, D and G: Alexa 488 or eGFP fluorescence. B, E and H: mRFP fluorescence. C, F and I: merged color images in which green corresponds to cathepsin A (panel C), saposin A (panel F) or eGFP-clathrin (panel I) and red corresponds to mRFP-hSR-BI. Scale bars = 10 µm. Representative images are shown.](/cms/asset/722822c9-9770-44e8-b52a-31af156f2471/imbc_a_229945_f0007_b.jpg)
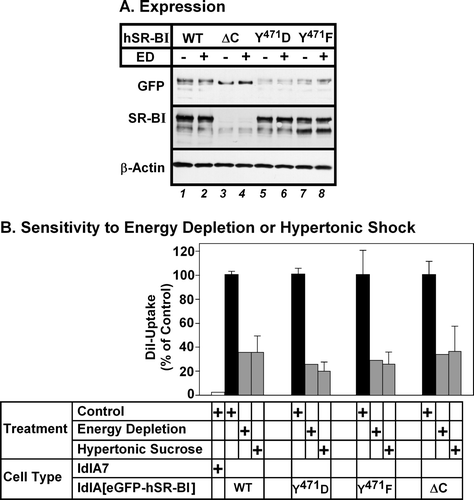
Figure 8. Mutations in the C-terminal cytoplasmic domain do not affect the dependence of hSR-BI mediated lipid uptake on endocytosis. LdlA7 cells were transfected with eGFP-hSR-BI (wild type, WT), eGFP-hSR-BI-Y471F, eGFP-hSR-BI-Y471D and eGFP-hSR-BI-ΔC expression plasmids and subjected to G418 selection. (A) Detergent extracts of control and energy depleted cells were prepared and proteins were subjected to SDS-PAGE and immunoblotting using antibodies specific for GFP (top panel), SR-BI-C terminal peptide (middle panel) and β-actin (loading control, bottom panel). Equal amounts of protein were applied to each lane. (B) Cells subjected to energy depletion or exposed to hypertonic sucrose (grey bars), control untreated cells (black bars) or untransfected ldlA7 cells (white bar) were incubated with DiI-HDL and DiI uptake and GFP-fluorescence were measured as described in the Methods section. The level of DiI uptake is expressed as the percentage of control, untreated cells and data represent the average±range of two independent experiments.