Figures & data
Figure 1. Membrane protein structures are hot. Analysis of high-resolution membrane protein structures collected by Stephen White (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html) indicates that novel structures (black bars) are often published in high-impact journals (i.e. Nature, Science or Cell) and accompanied by ‘high-profile accessories’ (i.e., commentaries and covers). Surprisingly, improved or follow-up structures (white bars) also maintain impact, albeit less than novel structures.
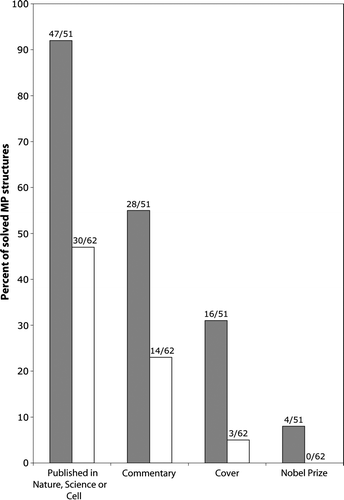
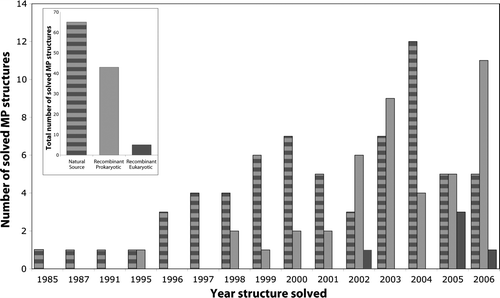
Figure 2. Eukaryotic membrane proteins produced by recombinant techniques are scarce in structural biology. Analysis of high resolution membrane protein structures collected by Stephen White (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html) indicates that most structures have been solved from proteins which have been purified from natural sources (striped bar), see inset. Of these, 44% were prokaryotic and 56% eukaryotic. Another successful approach is the production of recombinant prokaryotic proteins (light grey bar), see inset. In almost all of these cases the proteins have been produced in E. coli (not shown). Eukaryotic membrane proteins have not been successfully produced in the structural biology community to date (dark grey bar). Closer inspection indicates that most of the early structures were solved from proteins purified from natural sources, whereas recombinant technology arrived more recently. We are grateful to Niek Dekker (Astra Zeneca, Sweden) for assistance with this Figure.
Supplementary Table I. Structural status of the 25 largest MP families. Proteins collected by Stephen White (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html).
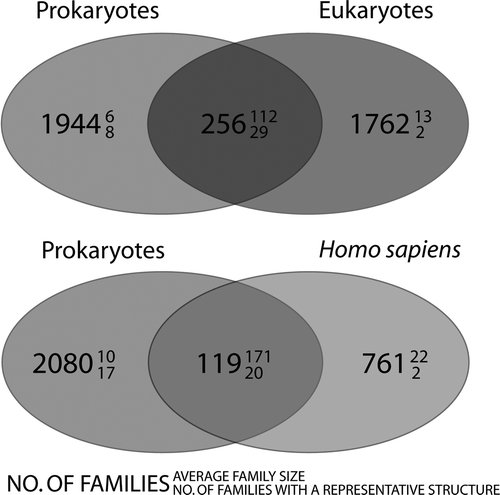
Figure 3. Venn diagrams, showing the distribution of membrane protein families. The upper panel shows the distribution of families between prokaryotes and eukaryotes and the lower panel shows the distribution of families between prokaryotes and Homo sapiens. The large number denotes the number of families that are common to that category. The number in superscript denotes the average family size and subscript, how many families in that category contain at least one member with known structure. It is evident that few are common to both prokaryotes and eukaryotes/Homo sapiens. These families are however very large (i.e., conserved across many species).