Figures & data
Table I. Primers for transcript analysis by semiquantitative RT PCR.
Table II. Amino acid sequence identity and average expression level of VHA subunits. (A) Comparison of amino acid sequence identity between the isoforms. The genes are identified by their At-number. (B, C) Average expression data are given as arithmetic mean and median, maximum and minimum values as calculated from 172 experiments. (B)% indicates the percent expression level compared to the total expression of this particular subunit. (C)%* indicates the percent expression of each subunit present without isoforms as compared to the average expression of all single isoform subunits.
Figure 1. Semi-quantification of subunit transcripts by RT-PCR and 2D-Western blot analysis. Results are shown for (A) shoots and (B) roots of plants grown under control (C) and salt stress (SS) conditions. Isoform transcripts were quantified and are depicted as relative percent ratios (A1, B1) or as median signal values (A2, B2). Representative PCR-results are also presented. The data are means of at least 4 RT PCRs from 2 independent experiments. Samples for 2D gel electrophoresis and Western blotting (A3, B3) were obtained from plants grown under control conditions. Western blot analysis was performed using antibody against AtVHA-E (1:3000), detection occurred via chemi-luminescence.
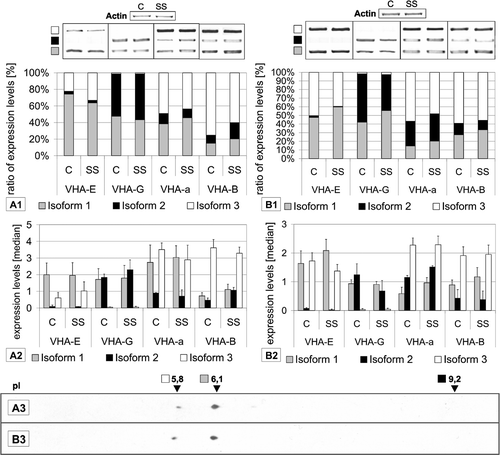
Figure 2. Tissue-specific expression of VHA isoforms. Expression levels were calculated as medians of signal values of the respective vha-genes using data from arrays hybridized with samples from the following tissues: leaves (l), roots (r), stems (st), petioles (pe), pollens (po), flowers (f), seedlings (se) and whole plant (wp). Diagrams on the left depict the ratio of expression levels of each isoform in percent of total expression level of all isoforms. In addition, the expression levels are presented as medians of signal values in order to indicate trends for absolute abundance.
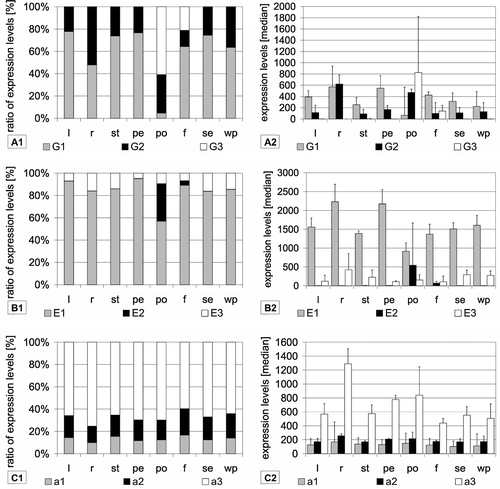
Figure 3. Stress specific expression of VHA-isoforms. Expression levels were calculated as medians of signal values. Data were used from the At-genexpress experiments performed under the following conditions: control (C), salt stress (SS), osmotic stress (OS), cold stress (CS), heat stress (HS) and drought stress (DS). Results are shown separately for experiments done with shoot tissue (A–C) and root tissue (D). Diagrams on the left present the ratio of expression levels of each isoform in percent of total expression level of all isoforms. In addition, the expression levels are presented as medians of signal values in order to present trends for absolute abundance.
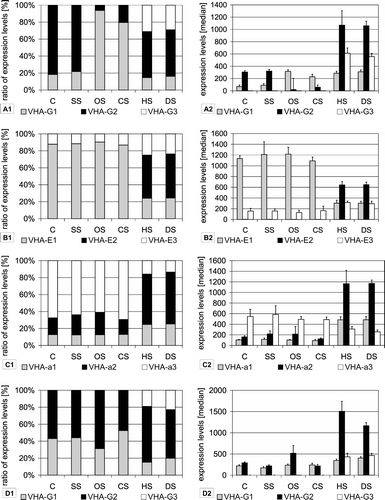
Figure 4. Correlation analysis of vha-gene expression among 172 experiments. For each subunit the signal values of all experiments were correlated pairwise with each subunit. The diagrams show only correlation factors (r) higher than 0.5, which point to a significant co-regulation of both genes. The correlation factor (r) values increase from the centre to the border of the diagram. (A) The correlation among all experiments (172). (B) Under stress conditions (85) and (C) for control experiments only (87).
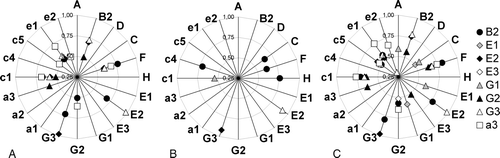
Figure 5. Amino acid sequence alignment of VHA-B subunits. Displayed is the alignment of VHA-B amino acid sequences from Neurospora crassa (Nc), Saccharomyces cereviseae (Sc), Homo sapiens (Hs), Oryza sativa (Os) and Arabidopsis thaliana (At). The alignment was generated with ClustalW.

Table III. Compilation of the rice VHA-genes. Each Arabidopsis thalianaVHA-cDNA was BLAST-searched against the rice genome or taken from the annotated TIGR rice genome database (db). Coding regions with significant similarity to A. thalianaVHA-genes were included in the table. VHA genes are given with Os-numbers from the TIGR db and the NCBI db. Additionally the number of amino acids (aa), the predicted molecular weight (MW) and the isoelectric point (pI) were calculated with the protparam tool. The identity of the rice sequences to A. thaliana was determined using ClustalW.