Figures & data
Figure 1. Analysis of the C-terminal domain of TRPV4. (A) Schematic drawing of the CFP, GFP, YFP and RFP-tagged TRPV4 constructs. (B) Various tagged sequential C-terminal deletion mutants were constructed (see Material and methods).
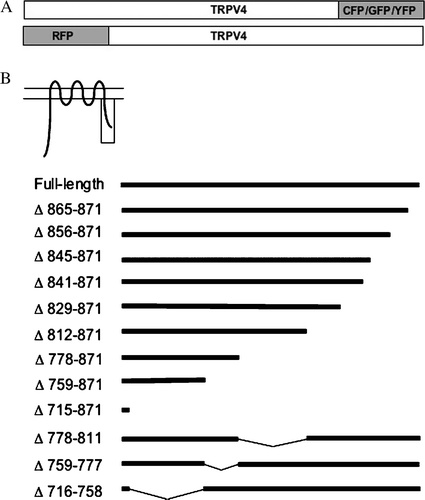
Figure 2. Full-length TRPV4 is localized in plasma membrane in different cell types. HaCaT keratinocytes (A) and CHO cells (B) were transiently transfected with TRPV4-GFP and fixed 48 h after transfection. The maximum intensity projections (MIP) taken with the laser scanning microscope (LSM), reveal the plasma membrane localization of TRPV4 in the different cell lines. TRPV4 is accumulated in lamellipodia and filopodia (arrowheads) as well as in microvilli (arrows). The orthogonal xz- and yz-sections (bottom and right) demonstrate clearly the localization of the channel in the plasma membrane in both cell types. In addition CHO cells display some TRPV4-GFP localization in the ER (indicated by the diffuse background fluorescence). Bar = 5 µm.
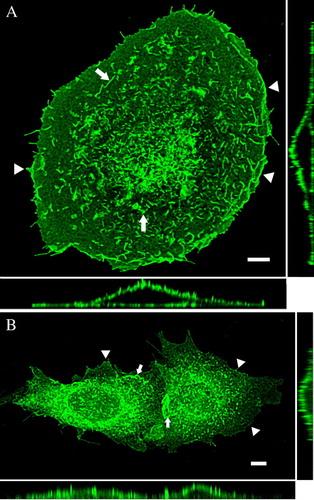
Figure 3. Differential localization of the deletion mutants: effect of the deletions and the cell type. HaCaT keratinocytes (A–D) and CHO cells (E–H) were transfected with the indicated deletion mutants, fixed after 48 h and analyzed with the LSM. (A) In HaCaT cells Δ856-871-GFP was completely localized in the plasma membrane (A, C), while Δ845-871 remained to a considerable amount in ER (B, D). The magnified insets clarify the exclusive plasma membrane localization of Δ856-871 (arrowheads) (C) and the assorted ER (arrows) and plasma membrane distribution (arrowheads) of Δ845-871 (D). In CHO cells Δ856-871 was mainly found in plasma membrane (arrowheads) (E and the magnified inset G), while Δ845-871 was exclusively localized in the ER (arrows) (F and the magnified inset H).
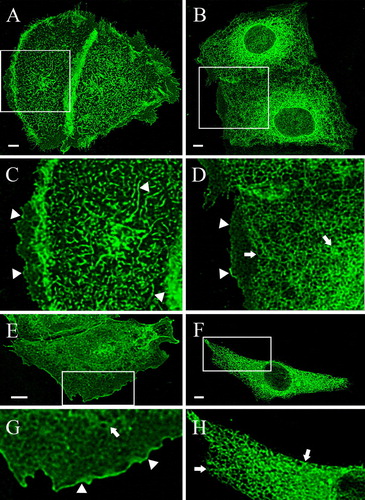
Figure 4. Cross complementation of deletion mutants with full-length TRPV4 rescues plasma membrane localization of Δ845-871, Δ841-871 and Δ829-871 in HaCaT keratinocytes. HaCaT keratinocytes were double transfected with full-length RFP-TRPV4 (red) and the GFP-tagged deletions mutants Δ856-871-GFP (A–C), Δ845-871-GFP (D–F), Δ841-871-GFP (G–I), Δ829-871-GFP (J–L) and Δ812-871-GFP (M–O) (green), fixed after 48 h and analyzed by LSM. The panels on the right display the merged images and the co-localization (yellow).
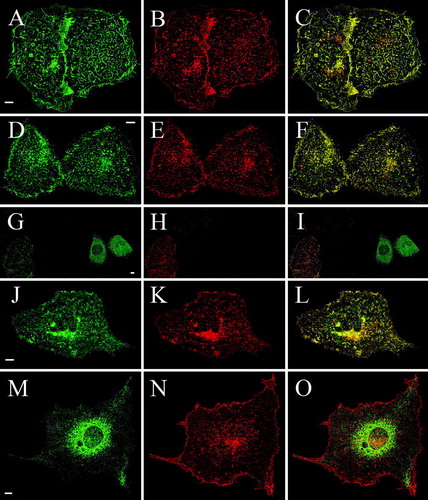
Table I. Expression patterns of TRPV4 deletion mutants after cotransfection with RFP-TRPV4 in HaCaT and CHO cells. HaCaT keratinocytes and CHO cells were double-transfected with the indicated GFP-tagged deletion mutants and RFP-TRPV4 and the localization of both recombinant proteins was analyzed by LSM. Coexpression of full-length RFP-TRPV4 with the deletion mutants Δ845-871-GFP, Δ841-871-GFP and Δ829-871-GFP (indicated in bold) showed a significant altered distribution in comparison with cells transfected with the respective mutant alone.
Table S1. Kinetics of TRPV4 expression in HaCaT keratinocytes and CHO cells. HaCaT keratinocytes and CHO cells were transfected with TRPV4-GFP and the localization of the recombinant protein was analyzed by LSM at the indicated time points. TRPV4-GFP in HaCaT cells is exported faster towards the plasma membrane than in CHO cells. Furthermore, TRPV4-GFP is HaCaT cells fully exported out of the ER in contrast to CHO cells.
Figure 5. Cross complementation of deletion mutants with full-length TRPV4 rescues plasma membrane localization of Δ845-871, Δ841-871 and Δ829-871 in CHO cells. CHO cells were double transfected with full-length RFP-TRPV4 (red) and Δ856-871-GFP (A–C), Δ845-871-GFP (D–F), Δ841-871-GFP (G–I), Δ829-871-GFP (J–L) and Δ812-871-GFP (M–O) (all green). Cells were fixed after 48 h and analyzed by LSM. On the right panels the merged images and the co-localization are displayed (yellow).
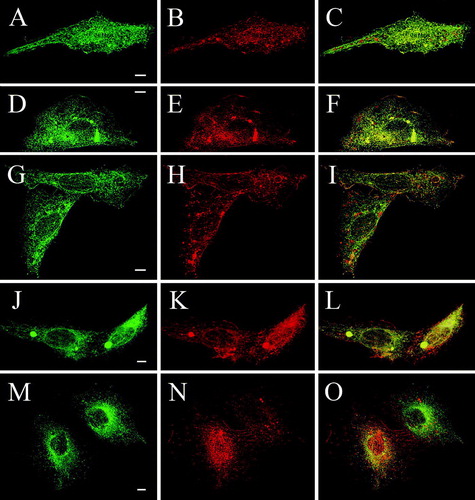
Figure 6. FRET of double transfected CHO cells. CHO cells were double transfected with TRPV4-CFP (A) and TRPV4-YFP (B) and fixed 48 h post transfection. The merged image (C) demonstrates the co-localization of the transfected constructs in the plasma membrane (PM) and the ER. (D) Acceptor bleaching in TRPV4-CFP and TRPV4-YFP as well as in TRPV4-CFP and Δ 841-871-YFP transfected cells exhibited a significantly higher FRET efficiency compared to TRPV4-CFP and Δ 812-871-YFP transfected cells in the PM and the ER. Bar = 10 µm.
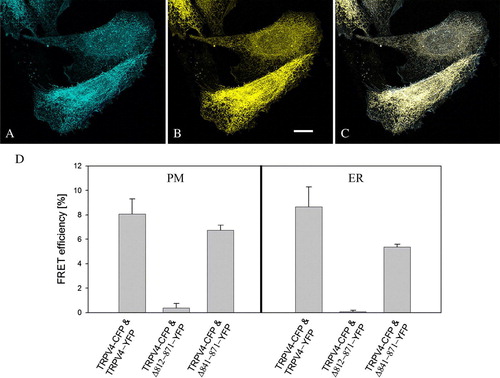
Figure 7. Deletion mutants are fully functional, when expressed in the plasma membrane. CHO cells were transiently transfected with different GFP deletion constructs, and after 48 h loaded with Fura-2 and exposed to hypotonicity by lowering the osmolarity from 300 to 200 mOsm/l. (A) In Δ841-871-GFP transfected cells, the [Ca2 + ]i stayed constant after hypotonic shock (n=27). In contrast [Ca2 + ]i was significantly increased in full-length TRPV4-GFP (n=14) as well as in Δ856-871-GFP transfected cells (n=21). (B) Representative traces of the three differently transfected cells were plotted. While the [Ca2 + ]i level remains constant in Δ841-871-GFP transfected cells (dotted line), Δ856-871-GFP (dashed line) and full-length TRPV4 (straight line) transfected cells reacted with a similar [Ca2 + ]i increase to the hypotonic stimulus (indicated by the line). Data demonstrated are mean values of±SEM.
![Figure 7. Deletion mutants are fully functional, when expressed in the plasma membrane. CHO cells were transiently transfected with different GFP deletion constructs, and after 48 h loaded with Fura-2 and exposed to hypotonicity by lowering the osmolarity from 300 to 200 mOsm/l. (A) In Δ841-871-GFP transfected cells, the [Ca2 + ]i stayed constant after hypotonic shock (n=27). In contrast [Ca2 + ]i was significantly increased in full-length TRPV4-GFP (n=14) as well as in Δ856-871-GFP transfected cells (n=21). (B) Representative traces of the three differently transfected cells were plotted. While the [Ca2 + ]i level remains constant in Δ841-871-GFP transfected cells (dotted line), Δ856-871-GFP (dashed line) and full-length TRPV4 (straight line) transfected cells reacted with a similar [Ca2 + ]i increase to the hypotonic stimulus (indicated by the line). Data demonstrated are mean values of±SEM.](/cms/asset/c87d70c5-7203-4b1a-ada0-61f2fb36a5c9/imbc_a_263369_f0007_b.gif)
Figure S1. Co-localization of TRPV4-GFP and a plasma membrane marker. HaCaT cells were transfected with TRPV4-GFP (green) and 48 h after transfection plasma membrane was counterstained using plasma membrane marker (see Materials and methods) (red). Nuclei were stained with Hoechst (blue). The four TRPV4-GFP transfected cells display strong co-localization with plasma membrane marker (yellow), demonstrating clearly TRPV4 localization in plasma membrane. Bar = 5 µm.
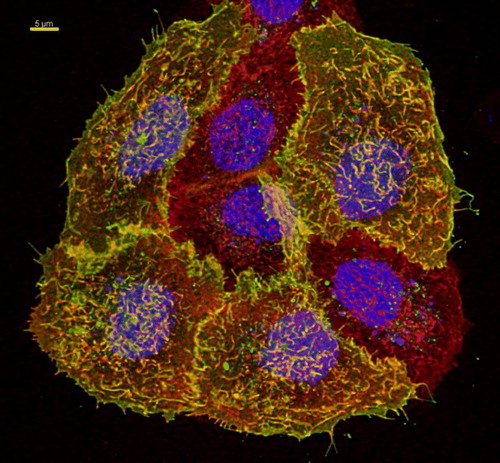
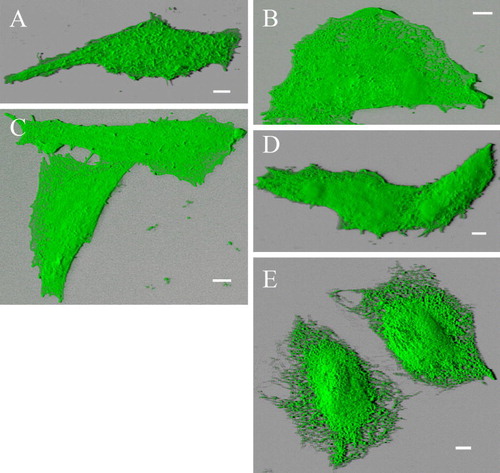
Figure S2. Blended 3D projection of the GFP channel reveals plasma membrane- and ER-localization of different deletion mutants. Alternative blended shadow projection reveals evidently plasma membrane localization of truncation mutants Δ856-871-GFP (A), Δ845-871-GFP (B), Δ841-871-GFP (C) and Δ829-871-GFP (D) double transfected with RFP-TRPV4 in CHO cells. In contrast Δ829-871-GFP (E) is exclusively localized in ER. Δ856-871-GFP transfected cell displayed more Microvilli structures than the other cells, resulting in rougher plasma membrane surface. Bar = 5 µm.