Figures & data
Figure 1. Identification of A. nidulans transformants expressing a mutant version of the human cac cDNA at physiological level. (A) Schematic representation of the integration at the chromosomal argB locus of a single-copy of plasmid pARGHCC that enables expression of hCAC with a defined mutation in its RX2PANAAXF motif. Chromosomal and plasmid EcoRI sites are indicated. The EcoRI fragment of A. nidulans PPA14 (recipient strain) and the two EcoRI fragments of argB+ single-copy transformants detected by the argB probe used are shown by arrows. Sizes of these fragments are also given. Mutation at the argB2 loci is indicated by an open box. (B) Southern blotting of genomic DNA from PPA14 (ΔacuH; argB2) (lane1) and argB+ transformants (lanes 2–9) digested with EcoRI and hybridized with an 1.7-kb HindIII internal fragment of the A. nidulans argB gene. In this blot, transformants in lanes 2, 3 and 9 contain a single-copy of the cac transgene that encodes the R275K mutant hCAC. (C) Confirmation that previously selected argB+ single-copy transformants contain one copy of the human cac transgene was performed by Southern blotting using the cac cDNA as a probe. In this analysis, the PPA14 strain (lane 1) was employed as negative control, whereas correct transformants (lanes 2–9) give a 7.5-kb hybridization band. (D) Wild-type and mutant hCACs are similarly expressed and localized in A. nidulans mitochondria, as confirmed by Western blotting. A. nidulans cells expressing wild-type hCAC (lane 1) or the R275K (lane 2), P278G (lane 3), A279G (lane 4), N280Q (lane 5), A281G (lane 6), A282G (lane 7) and F284G (lane 8) mutated versions were grown for 20 h on 0.3% sucrose + 0.1 M acetate minimal medium and used to obtain a 17000 g mitochondrial-enriched fraction. Proteins (20 µg) from this fraction were separated on SDS-PAGE, blotted onto nitrocellulose membranes and immunodetected with specific anti-CAC antibodies.
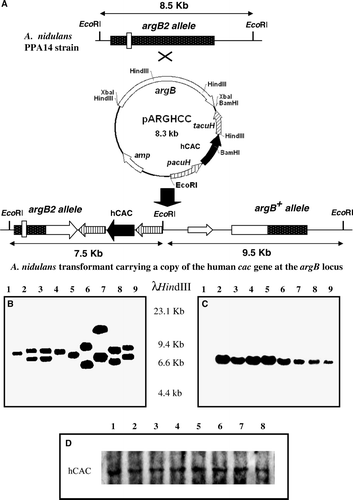
Figure 2. Growth analysis on glucose and acetate minimal medium of Aspergillus nidulans strains expressing different human CACs. A. nidulans single-copy transformants expressing the following versions of the hCAC: wild-type (hCAC) and R275K, P278G, A279G, N280Q, A281G, A282G, F284G mutants were streaked out on plates containing 1% glucose minimal medium (A) or 0.1 M acetate minimal medium (B) and incubated at 37°C for 3 days. The A. nidulans ΔacuHT14 (ΔACUH) strain was cultivated as a negative control. This Figure is reproduced in colour in Molecular Membrane Biology online.
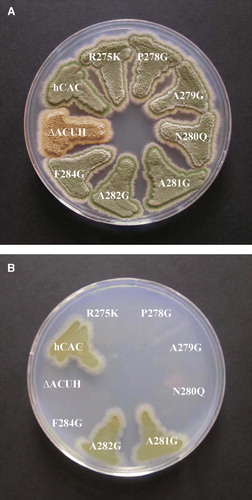
Figure 3. Quantification of functionality of mutant hCACs in A. nidulans by microcalorimetric analysis. (A) Power-time (p-t) curve on 0.1 M acetate minimal medium of A. nidulans PPA14 (ΔacuH; argB2) strain and single-copy transformants expressing the following versions of the hCAC at the physiological level: wild-type (hCAC), R275K, A281G, A282G mutants. Symbols representing each (p-t) curve are indicated at the bottom of the plot. (B) Semilogarithmic p-t representation during exponential growth of A. nidulans single-copy transformants expressing the wild-type (hCAC) and the A281G or A282G mutant versions. Plot symbols are indicated at the bottom.
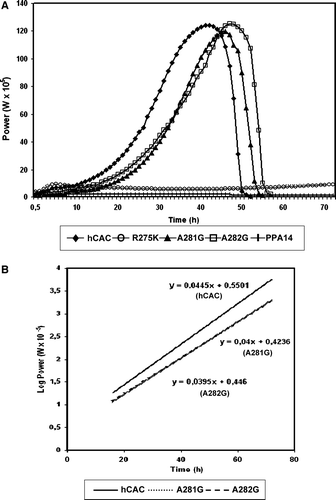
Figure 4. Time course of [3H]carnitine uptake by liposomes reconstituted with recombinant hCAC proteins. Transport was started by adding 0.1 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and stopped at the indicated times. In each panel the data relative to the wild-type hCAC (○) are shown with those relative to the mutants R275K (•) and R275A (□) in (A); P278G (•) and P278V (□) in (B); N278G (•) and N280Q (□) in (C); A279G (•), A281G (□), A282G (▪) and F284G (r) in (D). The data represent means±S.D. of three independent experiments carried out in duplicate.
![Figure 4. Time course of [3H]carnitine uptake by liposomes reconstituted with recombinant hCAC proteins. Transport was started by adding 0.1 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and stopped at the indicated times. In each panel the data relative to the wild-type hCAC (○) are shown with those relative to the mutants R275K (•) and R275A (□) in (A); P278G (•) and P278V (□) in (B); N278G (•) and N280Q (□) in (C); A279G (•), A281G (□), A282G (▪) and F284G (r) in (D). The data represent means±S.D. of three independent experiments carried out in duplicate.](/cms/asset/c229954c-78f2-4a8c-a02a-fe8eaebec993/imbc_a_269628_f0004_b.gif)
Table I. Kinetic constants of reconstituted wild-type and mutant hCACs. The values were calculated from double-reciprocal plots of the rates of carnitine/carnitine exchange under variation of the external substrate concentration. The exchange was started by the addition of 0.12–2.0 mM [3H]carnitine to proteoliposomes containing 13 mM carnitine and reconstituted with the wild-type hCAC or one of the indicated mutants. The reaction time was 4 min. The Km values of mutant carriers that are severely affected in their transport activity by the indicated mutation were not assayed (n.d., not determined). The data represent the means±SD of four different experiments.
Table II. Transport of acetylcarnitine or carnitine in exchange for [3H]carnitine by liposomes reconstituted with recombinant hCAC proteins. Proteoliposomes were preloaded internally with 13 mM acetylcarnitine or 13 mM carnitine. Transport was started by the external addition of 0.1 mM [3H]carnitine and terminated after 10 min. The data represent means±SD of three independent experiments.
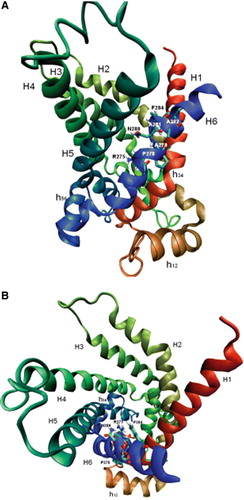
Figure 5. Structural model of the hCAC protein. Ribbon diagrams viewing the carrier from the lateral side (A) and the cytosolic side (B). The transmembrane α-helices are coloured as follows: H1 red, H2 orange-green, H3 light-green, H4 dark-green, H5 light-blue and H6 blue. Stick representation highlights some residues of the motif R-X-X-P-A-N-A-A-X-F indicated by their number in the hCAC sequence.