Figures & data
Figure 1. Constitutive SSY1 mutants have increased apparent affinity for leucine, phenylalanine and L-norvaline. Exponentially growing M5445 cells expressing SSY1 wild type or SSY1 constitutive alleles (SSY1-102, SSY1-130, SSY1-137, SSY1-150 and SSY1-151) were exposed for 10 min to leucine, phenylalanine or L-norvaline at various concentrations, and the EC50 for Stp1p processing was determined. Experiments were performed at least twice. SEM is shown by error-bars.
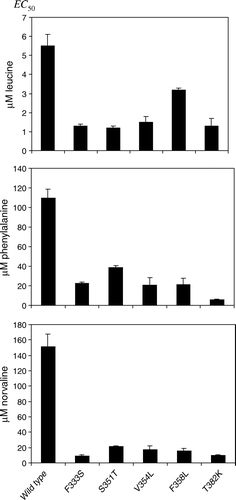
Figure 2. Dose-response analysis of SSY1 mutants. Exponentially growing M5445 cells expressing SSY1 wild type, SSY1-137 or ssy1-23 harbored on centromeric plasmids were induced with a range of concentrations of leucine, phenylalanine or L-norvaline. Stp1p processing was quantified, and the EC50 was estimated. Circles: M5445/pSSY1-137. Triangles: M5445/pSSY1. Squares: M5445/pSSY1-23.
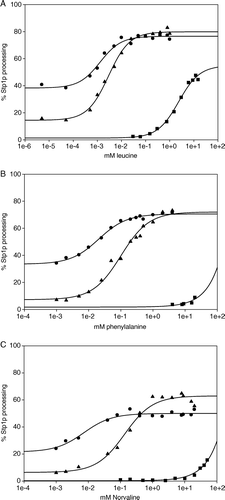
Table I. Basal Stp1p processing of SSY1 mutations expressed in M5445.
Table II. Constitutive SPS sensor components interact co-operatively to increase Stp1p processing.
Table SI. Strains used in this study.
Table SII. Primers used for introduction of site-specific mutations in SSY1.
Table SIII. Positions of mutations and corresponding amino acid substitutions in SSY1 mutants.
Figure 3. Amino acid sequence alignment of S. cerevisiae Ssy1p and A. aeolicus LeuTAa. The sequences were aligned using the FFAS03 algorithm Citation[28]. Transmembrane α-helices (TM1 through TM12) and an extracellular α-helix (EL2) of LeuTAa are highlighted in grey. Positions of single amino acid substitutions in Ssy1p that result in constitutive signaling are indicated by stars: F333S, S351T, V354L, F358L, T382K, V694F and G790V; substitution T639I, causing hypo-responsiveness, is also indicated by a star.
![Figure 3. Amino acid sequence alignment of S. cerevisiae Ssy1p and A. aeolicus LeuTAa. The sequences were aligned using the FFAS03 algorithm Citation[28]. Transmembrane α-helices (TM1 through TM12) and an extracellular α-helix (EL2) of LeuTAa are highlighted in grey. Positions of single amino acid substitutions in Ssy1p that result in constitutive signaling are indicated by stars: F333S, S351T, V354L, F358L, T382K, V694F and G790V; substitution T639I, causing hypo-responsiveness, is also indicated by a star.](/cms/asset/319b261c-3f0e-40f7-b98d-03e80179236c/imbc_a_277184_f0003_b.gif)
Figure 4. (A) Positions of amino acid substitutions that influence signaling by Ssy1p indicated in the secondary structure of LeuTAa. The activating substitutions F333S in TM2, S351T, V354L and F358L in TM3, T382K in EL3, V694F in TM9, and G790V in TM12 are indicated by green spheres, whereas the substitution T639I in TM8, which leads to hypo-responsiveness, is indicated by a red sphere. (B) Model of Ssy1p based on its similarity to LeuTAa. Modeling was carried out at (http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) using the Jackal modeling method and the All-atom model. As above, substitutions in Ssy1p are indicated by green and red spheres.
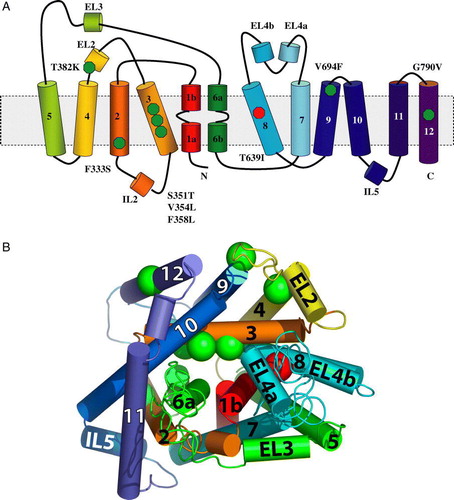
Figure 5. Model of the mechanism of amino acid signaling. In the absence of extracellular amino acids the plasma membrane receptor Ssy1p has a strong preference for an inward-facing conformation and is present in a complex with Ptr3p and the Ssy5p protease, which is inhibited by its interaction with the Ssy5p prodomain. Binding of an extracellular amino acid to Ssy1p stabilizes an altered conformation of the whole complex in which Ssy1p is outward-facing. This event results in the release of the inhibitory Ssy5p prodomain and processing of Stp1p. The figure is a simplified view, and does not consider how phosphorylation/dephosphorylation and ubiquitination Citation[15], Citation[16], Citation[36], Citation[37] may participate in signaling.
![Figure 5. Model of the mechanism of amino acid signaling. In the absence of extracellular amino acids the plasma membrane receptor Ssy1p has a strong preference for an inward-facing conformation and is present in a complex with Ptr3p and the Ssy5p protease, which is inhibited by its interaction with the Ssy5p prodomain. Binding of an extracellular amino acid to Ssy1p stabilizes an altered conformation of the whole complex in which Ssy1p is outward-facing. This event results in the release of the inhibitory Ssy5p prodomain and processing of Stp1p. The figure is a simplified view, and does not consider how phosphorylation/dephosphorylation and ubiquitination Citation[15], Citation[16], Citation[36], Citation[37] may participate in signaling.](/cms/asset/0eee0cef-1e3a-438f-bcd5-ec00a21df753/imbc_a_277184_f0005_b.jpg)
Figure S1. Representative immunoblots, forming the basis of dose-response relationships presented in of the article. Extracts from exponentially growing M5445 cells expressing SSY1 wild type, SSY1-137 or ssy1-23 harboured on centromeric plasmids were analysed. The processing of transcription factor Stp1p was monitored in response to induction with a range of concentrations of leucine and L-norvaline, using a chromosomally encoded, translational fusion (Stp1p-ZZ) in which a doublet of the IgG-binding Z domain of the Staphylococcus aureus protein A had been fused to the C-terminus of Stp1p as described (Poulsen et al. 2005b). The unprocessed and processed forms of Stp1p-ZZ are seen as the slow- and fast-migrating bands, respectively, differing in molecular mass by 10 kDa, the amount by which Stp1 is truncated in the signalling (Andréasson & Ljungdahl [Citation2002]). With SSY1 wild type and SSY1-137 the range of leucine concentrations was from 0.005 µM to 1000 µM, whereas 0.03 mM to 15 mM leucine was used with ssy1-23 (panel A, quantification shown in A of the article). L-Norvaline was used in a concentration range from 0.001 mM to 20 mM for SSY1 wild type and SSY1-137, whereas a range from 0.1 mM to 50 mM norvaline was used with ssy1-23 (panel B, quantification shown in C of the article).
![Figure S1. Representative immunoblots, forming the basis of dose-response relationships presented in Figure 2 of the article. Extracts from exponentially growing M5445 cells expressing SSY1 wild type, SSY1-137 or ssy1-23 harboured on centromeric plasmids were analysed. The processing of transcription factor Stp1p was monitored in response to induction with a range of concentrations of leucine and L-norvaline, using a chromosomally encoded, translational fusion (Stp1p-ZZ) in which a doublet of the IgG-binding Z domain of the Staphylococcus aureus protein A had been fused to the C-terminus of Stp1p as described (Poulsen et al. 2005b). The unprocessed and processed forms of Stp1p-ZZ are seen as the slow- and fast-migrating bands, respectively, differing in molecular mass by 10 kDa, the amount by which Stp1 is truncated in the signalling (Andréasson & Ljungdahl [Citation2002]). With SSY1 wild type and SSY1-137 the range of leucine concentrations was from 0.005 µM to 1000 µM, whereas 0.03 mM to 15 mM leucine was used with ssy1-23 (panel A, quantification shown in Figure 2A of the article). L-Norvaline was used in a concentration range from 0.001 mM to 20 mM for SSY1 wild type and SSY1-137, whereas a range from 0.1 mM to 50 mM norvaline was used with ssy1-23 (panel B, quantification shown in Figure 2C of the article).](/cms/asset/a0939815-6345-44de-91df-edecbe860faa/imbc_a_277184_f0006_b.gif)
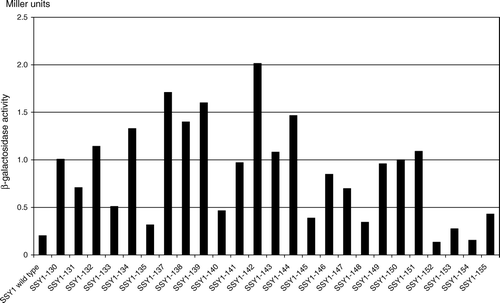
Figure S2. SSY1 mutations that cause constitutive activation of the AGP1 promoter. β-Galactosidase activity in extracts of strain M5380 (ssy1Δ AGP1::PAGP1-lacZ) transformed with centromeric plasmids expressing wild type SSY1 or the mutant SSY1 alleles obtained in screen A (SSY1-130 through SSY137), screen B (SSY1-138 through SSY1-149) or by site-directed mutagenesis (SSY1-150 through SSY155). The -galactosidase activities are the means of at least two independent determinations with less than 20% deviation.