Figures & data
Figure 1. Schematic representation of the proteins used in this work. The precursor of SphB1 is shown on the upper line, with SP, S, X and β representing the signal peptide, subtilase domain, ‘X’ domain and predicted β-barrel domain, respectively (approximately drawn to scale). The white boxes represent linker regions between the various domains, without well-predicted secondary structures. The hatched box represents a long predicted α-helix most likely corresponding to that of NalP. The arrowhead indicates the natural maturation site, and the sequence below corresponds to the N-terminus of the translocator in the outer membrane. The truncated versions of SphB1 were produced with an E. coli signal peptide (not represented) followed by an affinity Strep-tag (black box). The sequences underneath the various derivatives indicate the first residues of the SphB1 portion in each construct. The vertical arrows indicate the identified trypsin cleavage site in SphB1-X-α-β and SphB1-α-β. Its N-terminal sequence corresponds to residues 3–9 of the N-terminus of the translocator domain naturally found in the outer membrane of B. pertussis and arising from autoproteolytic cleavage of the protein.
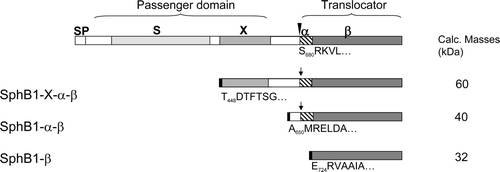
Figure 2. Folding state of purified SphB1-X-α-β and SphB1-α-β proteins. (A) Heat modifiability and trypsic digestion of the purified proteins. Lanes 1 and 3: purified SphB1-X-α-β and SphB1-α-β, respectively. The samples were heated for 10 min at 95°C. Lanes 2 and 4: SphB1-X-α-β and SphB1-α-β, non-heated samples. The proteins were subjected to SDS-PAGE, and the gels were coloured with Coomassie Blue. (B) CD spectra of SphB1 derivatives. Solid line, SphB1-α-β; dashed line, SphB1-X-α-β.
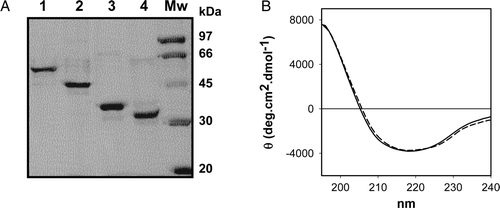
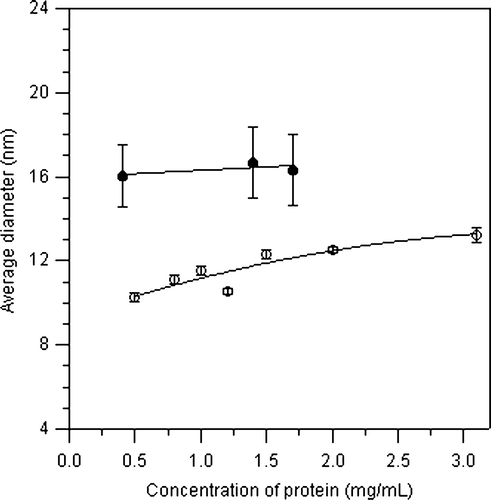
Figure 3. Dynamic light scattering analysis of the SphB1 recombinant proteins. (A and B) Volume size distribution of SphB1-α-β/DDM (solid line) and SphB1-X-α-β/DDM (dashed line) micelles without (A) and with 1% SDS (B). (C) Variation of the average diameter of SphB1-α-β/DDM (open symbols) and SphB1-X-α-β/DDM (filled symbols) micelles as a function of the SDS concentration. The lines are drawn as guides for the eye. The concentration of the protein is 1.2 mg/mL and 1.4 mg/mL for SphB1-α-β and SphB1-X-α-β, respectively.
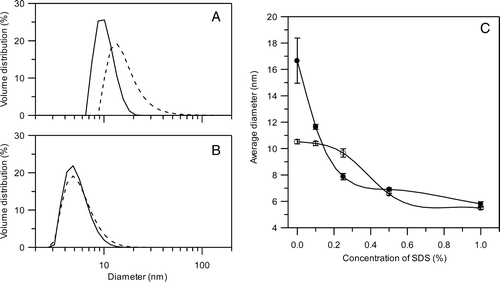
Figure 4. Cross-linking of purified SphB1-X-α-β and SphB1-α-β. The proteins were incubated for 1 h with the DSS cross-linker at 2 mM. Lanes 1–4: SphB1-α-β. Lanes 5–8: SphB1-X-α-β. ‘+’ and ‘−’ indicate the addition of cross-linker to the proteins and no addition, respectively. Lanes 2 and 6: the cross-linking reaction was performed in the absence of SDS, while lanes 3 and 7 and lanes 4 and 8 correspond to cross-linking experiments performed in the presence of 0.1% and 1% of SDS, respectively. ‘*’ labelled the presumed dimers. The 12.5% SDS-PAGE gel was stained with Coomassie blue.
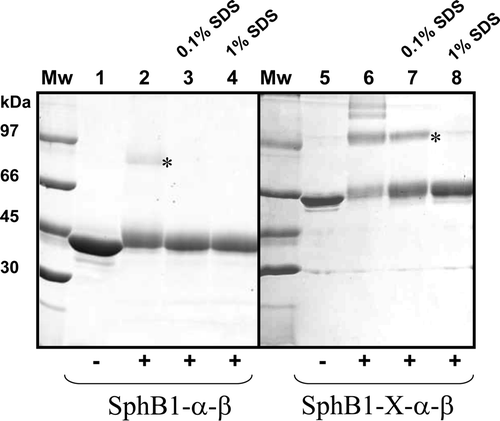
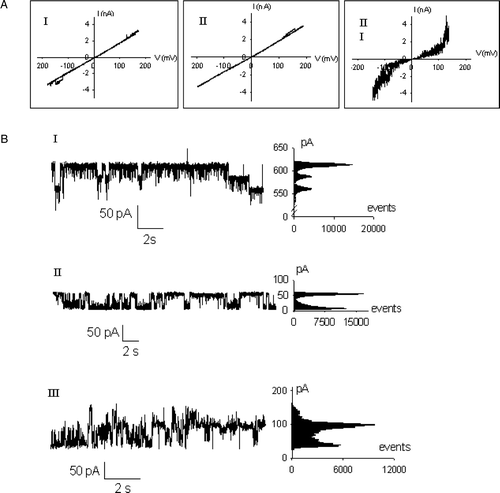
Figure 5. Channel activity of the SphB1 recombinant proteins. (A) Current /voltage recordings, with triangular ramps applied at a rate of 10 mV/sec to azolectin planar lipid bilayers containing around 0.5 µg of purified derivatives. (I) SphB1-X-α-β; (II) SphB1-α-β; (III) SphB1-β. (B) Current traces from single channels and associated amplitude histograms. (I) SphB1-X-α-β, applied voltage: 160 mV; (II) SphB1-α-β, applied voltage: 60 mV; (III) SphB1-β, applied voltage: 20 mV. The buffer used was 1 M KCl, 10 mM Hepes, pH 7.4.