Figures & data
Table I. Examples of peroxisomal function in mammals, yeast and plants.
Figure 1. PTS1 and PTS2 receptor recognition in various organisms. (A) In Arabidopsis PEX5 is required for both PTS1 and PTS2 mediated import. (B) In mammals two splice variants of PEX5 exist, a short isoform, PEX5S and a long isoform, PEX5L. PEX5L is required with PEX7 for PTS2 mediated import. Note: Both PEX5L and PEX5S function as the PTS1 receptor. (C) In S. cerevisiae the co-receptors PEX18 and PEX21 function with PEX7 for PTS2 import. (D) In N. crassa, P. pastoris, Y. lipolytica and H. polymorpha the co-receptor PEX20 functions with PEX7 for PTS2 import. PEX5 is not involved in PTS2 mediated import in yeast. In addition to receptor recognition, molecular chaperones may be required during import. These include members of the Hsp70-family Citation[115], Citation[116] and DnaJ-like proteins Citation[117], although the exact role of these chaperones remains to be determined. This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 1. PTS1 and PTS2 receptor recognition in various organisms. (A) In Arabidopsis PEX5 is required for both PTS1 and PTS2 mediated import. (B) In mammals two splice variants of PEX5 exist, a short isoform, PEX5S and a long isoform, PEX5L. PEX5L is required with PEX7 for PTS2 mediated import. Note: Both PEX5L and PEX5S function as the PTS1 receptor. (C) In S. cerevisiae the co-receptors PEX18 and PEX21 function with PEX7 for PTS2 import. (D) In N. crassa, P. pastoris, Y. lipolytica and H. polymorpha the co-receptor PEX20 functions with PEX7 for PTS2 import. PEX5 is not involved in PTS2 mediated import in yeast. In addition to receptor recognition, molecular chaperones may be required during import. These include members of the Hsp70-family Citation[115], Citation[116] and DnaJ-like proteins Citation[117], although the exact role of these chaperones remains to be determined. This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/c7fbf622-c2ee-4396-97c2-b2835c7bda2e/imbc_a_313224_f0001_b.jpg)
Figure 2. Overview of peroxisomal matrix protein import. Matrix proteins have either a PTS1 or PTS2 targeting signal that binds to a cytosolic receptor, PEX5 and PEX7 respectively (with or without co-receptors, see ) which targets the protein to the docking complex (PEX13, PEX14 and also PEX17 in yeast). The receptors translocate with their cargo into the peroxisome lumen, release their cargo and are recycled back into the cytosol. The RING-finger complex (PEX2, PEX10 and PEX12) associates with the docking complex via PEX8 (in yeast) and is required for receptor recycling. PEX10 is linked to the E2 ubiquitin conjugating enzyme, PEX4, which is anchored to the membrane by PEX22. PEX5 release from the membrane requires PEX4-dependent monoubiquitination, which is ATP-dependent. The RING-finger peroxins are the putative E3 ligases for this process. The AAA-proteins PEX1 and PEX6 are also involved in receptor release and are attached to the peroxisomal membrane via PEX15 in yeast and PEX26 in mammals. Little is known about the recycling of PEX7, although two of its co-receptors, PEX18 and PEX20, have been shown to be ubiquitinated. This Figure is reproduced in colour in Molecular Membrane Biology online.
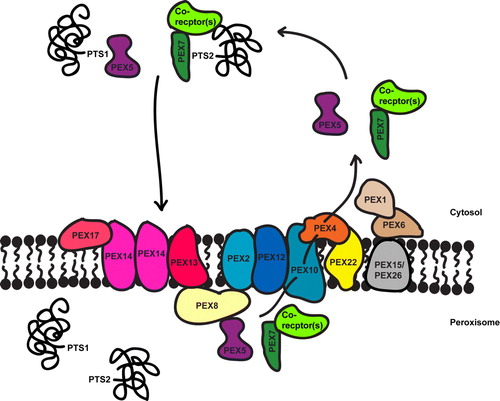
Figure 3. Model of PEX5 and PEX7 ubiquitination in receptor recycling and degradation. PEX5 recognizes PTS1 proteins in the cytosol and brings them to the docking complex (not shown). The receptor-cargo complex translocates to the luminal side of the peroxisomal membrane and disassociates, freeing the cargo. The receptor is then returned to the cytosol, likely through the monoubiquitination of PEX5 mediated by the UBC PEX4 and the putative E3 ligases PEX10/PEX2/PEX12 (RING). The AAA peroxins, PEX1 and PEX6 have been proposed to recognize the monoubiquitinated PEX5 and facilitate in its release from the membrane. PEX5 has also been shown to be polyubiquitinated by Ubc4p (and Ubc5p), although the E3 ligase responsible for polyubiquitination has not yet been determined. Polyubiquitinated PEX5 may again be recognized by the AAA peroxins and directed to the proteasome for degradation as a part of a quality control system. Asterisks = ubiquitin. Redrawn with permission from Citation[93]. This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 3. Model of PEX5 and PEX7 ubiquitination in receptor recycling and degradation. PEX5 recognizes PTS1 proteins in the cytosol and brings them to the docking complex (not shown). The receptor-cargo complex translocates to the luminal side of the peroxisomal membrane and disassociates, freeing the cargo. The receptor is then returned to the cytosol, likely through the monoubiquitination of PEX5 mediated by the UBC PEX4 and the putative E3 ligases PEX10/PEX2/PEX12 (RING). The AAA peroxins, PEX1 and PEX6 have been proposed to recognize the monoubiquitinated PEX5 and facilitate in its release from the membrane. PEX5 has also been shown to be polyubiquitinated by Ubc4p (and Ubc5p), although the E3 ligase responsible for polyubiquitination has not yet been determined. Polyubiquitinated PEX5 may again be recognized by the AAA peroxins and directed to the proteasome for degradation as a part of a quality control system. Asterisks = ubiquitin. Redrawn with permission from Citation[93]. This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/dc64cb29-af4d-4f79-8081-c76f79a4bcd8/imbc_a_313224_f0003_b.jpg)