Figures & data
Figure 1. Effect of increasing concentrations of different phospholipids on ACA8 hydrolytic activity. Assays were performed on ER enriched fraction from S. cerevisiae expressing ACA8 in presence of 40 µM free Ca2 + . The effect of phospholipids is expressed as percentage of stimulus relative to the activity measured in absence of added phospholipids (82 nmol Pi min−1 mg−1 protein). Results are from one experiment representative of at least three giving similar results. Assays were performed on three replicates and SE of the assay did not exceed 7% of the reported values.
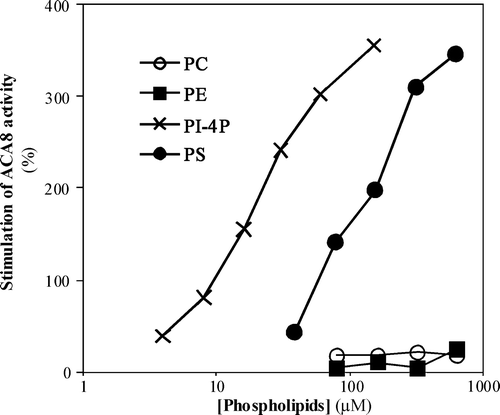
Figure 2. Effect of PS on ACA8 hydrolytic activity as a function of free Ca2 + concentration. Assays were performed on ER enriched fraction from S. cerevisiae expressing ACA8 in the presence (squares) or absence (circles) of 600 µM PS and in the presence (open symbols) or absence (closed symbols) of 1 µM CaM. Free Ca2 + concentration was buffered at 2.5–40 µM with 1 mM EGTA. Results (±SE, n=3) are from one experiment representative of three.
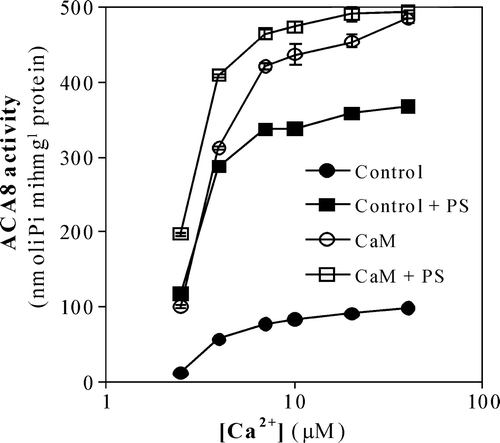
Table I. Effect of PS on the kinetic parameters of wild type ACA8 and of Δ74-ACA8 mutant measured in absence and in presence of 1 µM CaM. Values of Vmax and apparent K0.5 for free Ca2 + were evaluated by regression analysis of Ca2 + -ATPase activity measured as a function of free Ca2 + concentration as in ; r2>0.96. Results (±SE) are from three independent experiments.
Figure 3. Binding of phospholipids to ACA8 N-terminus. Different phospholipids (50 and 250 pmol) were spotted onto a nitrocellulose membrane which was then incubated with or without purified His- or GST-tagged N-terminus of ACA8 (6His-1M-I116 and GST-1M-I116) as described in the Experimental Procedures. Bound protein was visualized by immunodecoration with an antiserum directed against the portion 17V-T31 of ACA8. Results are from one experiment representative of five.
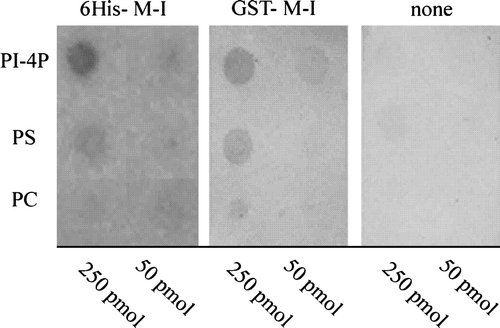
Figure 4. Effect of increasing concentrations of PI-4P on the activity of single point ACA8 mutants. Assays were performed on microsomal fraction from S. cerevisiae expressing wild type or single point ACA8 mutants in the presence of 10 µM free Ca2 + . Enzyme activation (f/fmax) is expressed as the ratio between PI-4P-dependent stimulus of Ca2 + -ATPase activity at the indicated PI-4P concentration and maximal stimulation (fmax) evaluated at saturating PI-4P concentration. Values of fmax ranged between 56% for the most deregulated mutant (R58A) (14) and 383% (wild type). Results are from one experiment representative of three giving similar results. Assays were performed on three replicates and SE of the assay did not exceed 5% of the reported values.
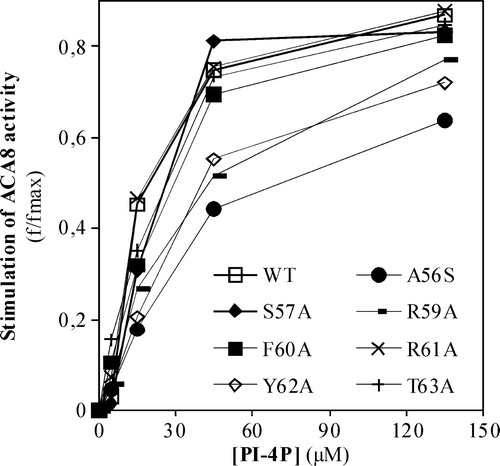
Figure 5. Effect of increasing concentrations of PS (panel A) or PI-4P (panel B) on the activity of N-deleted mutant Δ74-ACA8. Assays were performed on ER enriched fraction from S. cerevisiae expressing wild type or Δ74-ACA8 in the presence of 5 µM free Ca2 + and in the presence or in the absence of 1 µM CaM. Enzyme activation (f/fmax) is expressed as the ratio between phospholipid-dependent stimulus of Ca2 + -ATPase activity at the indicated phospholipid concentration and maximal stimulation (fmax) evaluated at saturating phospholipid concentration. Values of fmax of wild type, wild type in presence of CaM and Δ74-ACA8 were respectively 637%, 29% and 72% (PS, panel A) and 547%, 39% and 67% (PI-4P, panel B). Results (±SE, n=3) are from one experiment representative of three.
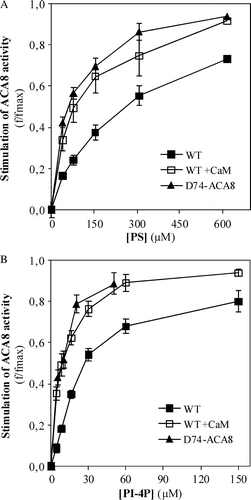
Figure 6. Helical wheel projection of ACA8 CaM-binding domain. A helical wheel projection of aa stretch R43-K68 of ACA8 was made using the program Binding Site Analysis in The Calmodulin Target Data Base. Residues involved in phospholipid binding are shown in bold; residues involved in auto-inhibition are underlined and those important in CaM recognition are in grey.