Figures & data
Supplementary Table 1. Oligonucleotides used in this study.
Figure 1. Relationships between So_ACR3 and other members of the ACR3 and BASS transporter families. The phylogenetic tree was constructed from the sequence alignment shown in Supplementary (online version only), using the Neighbour Joining method of Saitou and Nei [31]. The ACR3 proteins shown are representative of five subfamilies (i–v) identified by phylogenetic analysis of 334 ACR3 sequences obtained from the UniRef90 database [32].
![Figure 1. Relationships between So_ACR3 and other members of the ACR3 and BASS transporter families. The phylogenetic tree was constructed from the sequence alignment shown in Supplementary Figure 1 (online version only), using the Neighbour Joining method of Saitou and Nei [31]. The ACR3 proteins shown are representative of five subfamilies (i–v) identified by phylogenetic analysis of 334 ACR3 sequences obtained from the UniRef90 database [32].](/cms/asset/28a9ba38-527a-40fd-a323-6dfe43f6e604/imbc_a_353761_f0001_b.gif)
Figure 2. Sensitivity to arsenite and arsenate of E. coli AW3110 expressing wild-type So_ACR3, its P190A mutant or non-recombinant expression vector. Cultures were performed as detailed in Materials and methods. (A) Western blot of samples from cultures harbouring expression vectors encoding the indicated wild-type or P190A mutant forms of So_ACR3 in the presence or absence of 40 mM arsenate. Blots were stained with an antibody against oligohistidine. (B) and (C), growth of cultures, as monitored by D600nm, in the presence of the indicated concentrations of arsenite (As[III]) and arsenate (As[V]), respectively. The cultures were of bacteria expressing wild-type So_ACR3 (○), its P190A mutant (□), or harbouring non-recombinant expression vector (•).
![Figure 2. Sensitivity to arsenite and arsenate of E. coli AW3110 expressing wild-type So_ACR3, its P190A mutant or non-recombinant expression vector. Cultures were performed as detailed in Materials and methods. (A) Western blot of samples from cultures harbouring expression vectors encoding the indicated wild-type or P190A mutant forms of So_ACR3 in the presence or absence of 40 mM arsenate. Blots were stained with an antibody against oligohistidine. (B) and (C), growth of cultures, as monitored by D600nm, in the presence of the indicated concentrations of arsenite (As[III]) and arsenate (As[V]), respectively. The cultures were of bacteria expressing wild-type So_ACR3 (○), its P190A mutant (□), or harbouring non-recombinant expression vector (•).](/cms/asset/640bd6e4-5883-4530-9d5c-f8c88ac05825/imbc_a_353761_f0002_b.gif)
Figure 3. Comparison of wild-type So_ACR3 and its P190A mutant. (A) Imperial™-stained SDS-polyacrylamide gel of the purified proteins. The mobilities of marker proteins of known molecular mass are shown on the left. (B) Amide I region of the FTIR spectra of hydrated films of the wild-type and mutant proteins and bands obtained by deconvolution (solid lines). The latter were assigned to β-sheet (i and vi), unordered structure (ii), α-helix (iii and iv) and β-turns (v, va and vb). The curves fitted using these component bands are shown as dotted lines.
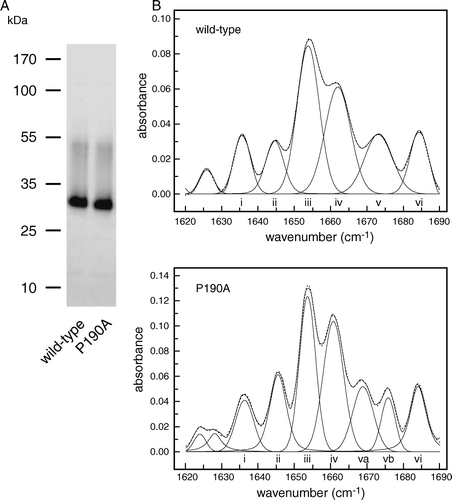
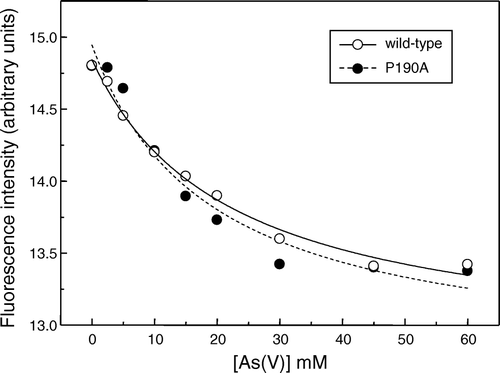
Figure 4. Concentration-dependence of the quenching by arsenate of the intrinsic fluorescence of wild-type So_ACR3 (○) and its P190A mutant (•). The fluorescence of samples of the purified proteins was measured in the presence of the indicated arsenate concentrations. The curves represent non-linear fits to the equation of a hyperbola, as detailed in the text.
Supplementary Table 2. Analysis of the secondary structure of wild-type and mutant So_ACR3 by FTIR spectroscopy.
Figure 5. Accessibility to membrane-impermeant thiol reagents of a cysteine residue introduced at position 3 in So_ACR3. Intact cells expressing a C126A/S3C double mutant of So_ACR3 were incubated directly with OGM (lane 1), or incubated with (lane 3) or without (lane 2) MTSET before isolation of membranes and subsequent incubation with OGM as detailed in Materials and Methods. Following affinity-purification of So_ACR3, samples were subjected to SDS-PAGE followed by detection of protein fluorescence (panel on the right) and subsequent staining for protein (panel on the left).
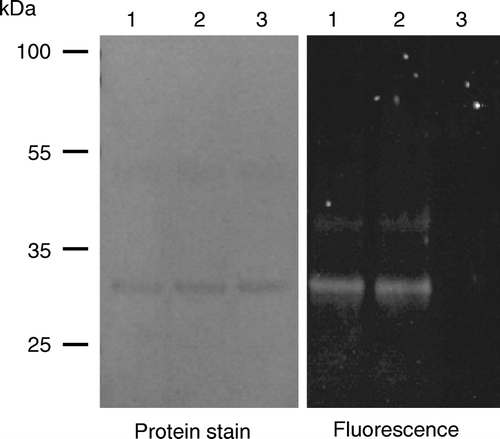
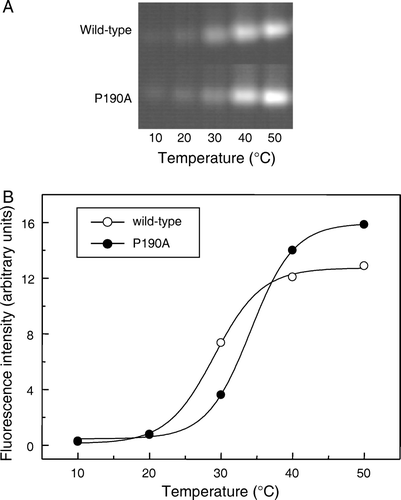
Figure 6. Effect of temperature on the reactivity of wild-type and mutant So_ACR3 towards OGM. Following incubation of the purified proteins with OGM at the indicated temperatures, samples were resolved by SDS-PAGE and fluorescence visualized (A). Panel B shows the incorporation of fluorescent label, quantified as described in Materials and methods, into wild-type So_ACR3 (○) and its P190A mutant (•). The curves represent non-linear fits of the data to a Boltzmann sigmoidal equation, performed in order to estimate apparent Tm values.
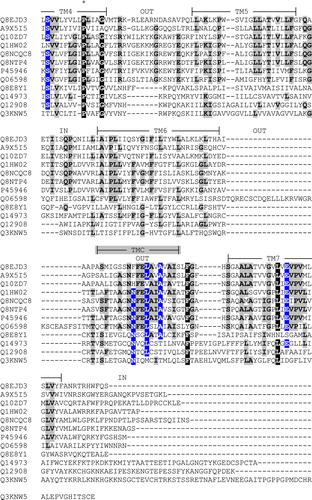
Supplementary Figure 1. Alignment of the amino acid sequences of So_ACR3 and other members of the ACR and BASS transporter families. Sequences are identified by their UniProt accession numbers as follows: Q8EJD3, So_ACR3 from Shewanella oneidensis MR-1; A9X5I5, Acr3 from Ochrobactrum tritici; Q10ZD7, putative arsenical resistance protein from Trichodesmium erythraeum; Q1HW02, ArsB from Streptomyces sp. FR-008; Q8NCQC8, ArsB1 from Corynebacterium glutamicum; Q8NTP4, ArsB2 from C. glutamicum; P45946, ArsB/Acr3 from Bacillus subtilis; Q06598, ACR3 from Saccharomyces cerevisiae; Q8E8Y1, putative bile salt transporter from S. oneidensis MR-1; Q14973, SLC10A1/NTCP from Homo sapiens; Q12908, SLC10A2/ASBT from H. sapiens; Q3KNW5, SLC10A6/SOAT from H. sapiens. The alignment was created using the program MUSCLE [1] followed by manual adjustment where necessary. X, Y and Z respectively indicate residues that are identical in 6–8, in 9–10 and in 11–12 of the 12 sequences. The experimentally determined topology and predicted locations of seven transmembrane segments (TM1-7) in human SLC10A2 shown on the Figure are taken from the work of Banerjee and Swan [2]. The indicated locations of three additional, putative transmembrane regions in ArsB/Acr3 from Bacillus subtilis (TMA-C) are taken from the work of Altonen and Silow [3]. An asterisk (*) shows the location of So_ACR3 residue proline 190.
![Supplementary Figure 1. Alignment of the amino acid sequences of So_ACR3 and other members of the ACR and BASS transporter families. Sequences are identified by their UniProt accession numbers as follows: Q8EJD3, So_ACR3 from Shewanella oneidensis MR-1; A9X5I5, Acr3 from Ochrobactrum tritici; Q10ZD7, putative arsenical resistance protein from Trichodesmium erythraeum; Q1HW02, ArsB from Streptomyces sp. FR-008; Q8NCQC8, ArsB1 from Corynebacterium glutamicum; Q8NTP4, ArsB2 from C. glutamicum; P45946, ArsB/Acr3 from Bacillus subtilis; Q06598, ACR3 from Saccharomyces cerevisiae; Q8E8Y1, putative bile salt transporter from S. oneidensis MR-1; Q14973, SLC10A1/NTCP from Homo sapiens; Q12908, SLC10A2/ASBT from H. sapiens; Q3KNW5, SLC10A6/SOAT from H. sapiens. The alignment was created using the program MUSCLE [1] followed by manual adjustment where necessary. X, Y and Z respectively indicate residues that are identical in 6–8, in 9–10 and in 11–12 of the 12 sequences. The experimentally determined topology and predicted locations of seven transmembrane segments (TM1-7) in human SLC10A2 shown on the Figure are taken from the work of Banerjee and Swan [2]. The indicated locations of three additional, putative transmembrane regions in ArsB/Acr3 from Bacillus subtilis (TMA-C) are taken from the work of Altonen and Silow [3]. An asterisk (*) shows the location of So_ACR3 residue proline 190.](/cms/asset/00343646-474b-49bf-8e10-a37051a1890a/imbc_a_353761_f0007_b.jpg)