Figures & data
Figure 1. Localization of lectin PNA by fluorescence microscopy. (A) A distinct staining of the brush border surface is seen along the villi. In addition, goblet cells (marked by an arrow) appear as strongly labeled dots. (B) A higher magnification image of the villus shows strong labeling of the secretory granules of goblet cells. Bars: 25 µm (A), 15 µm (B). Published in colour in the online version.
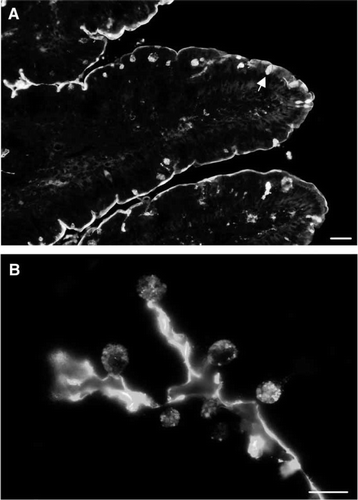
Figure 2. Identification of galectin-2 in a DRF prepared from microvillar membranes. A microvillar DRF was prepared as described in Methods, resuspended in 25 mM HEPES-HCL, 150 mM NaCl, pH 7.1, and incubated for 30 min at room temperature in the absence or presence of 0.1 M lactose. After incubation, the DRF was centrifuged at 20,000 g for 30 min, and the pellets (in three different loadings) were analyzed by SDS-PAGE, followed by transfer onto an immobilon PVDF membrane. Protein staining revealed a distinct lactose-sensitive band at ∼14 kDa. This band was carefully excised from the gel track furthest to the left and identified by MALDI-TOF analysis as galectin-2. The identification was verified by immunoblotting with an antibody to galectin-2. (The part of the membrane excised for MALDI-TOFF analysis is marked by an asterisk on the immunoblot image). Molecular weight values are indicated by arrows.
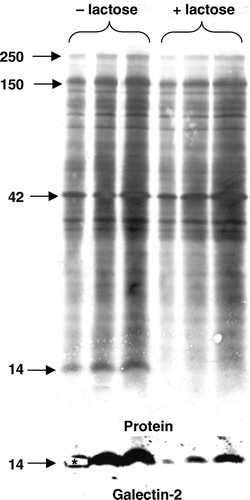
Figure 3. Subcellular localization of galectin-2. (A) SDS-PAGE of Mg2+-precipitated (basolateral and intracellular) membranes (Mg), soluble proteins (Sol), microvillar membranes (Mic), and DRF prepared from jejunal mucosa, as described in Methods. After electrophoresis, alkaline phosphatase (AP, 67 kDa) and galectin-2 (Gal-2) were visualized by immunoblotting and total protein by staining with Coomassie Brilliant Blue. Molecular weight values are indicated by arrows. (B) Samples of a right-side-out microvillar membrane vesicle fraction were resuspended in 25 mM HEPES-HCl, 150 mM NaCl, pH 7.1, containing 0 mM, 10 mM or 50 mM of lactose, respectively. After incubation for 30 min at room temperature the samples were centrifuged at 20,000 g for 30 min. The pellets and supernatants were subjected to SDS/PAGE, followed by immunoblotting with an antibody to galectin-2.
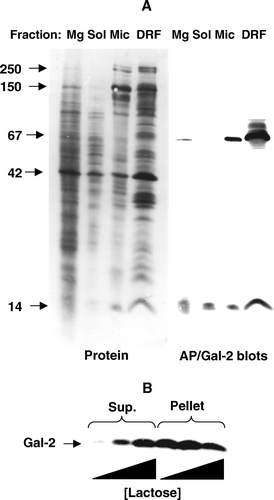
Figure 4. Localization of galectin-2 and galectin-4 in the jejunum by immunofluorescence microscopy. Double labeling for galectin-2 and galectin-4 on paraffin sections was performed as described in Methods. Labeling for both galectins is seen along the brush border of the villi. Extensive, but not complete colocalisation of the two galectins is evident in the merged micrograph. Bars, 25 µm. Published in colour in the online version.
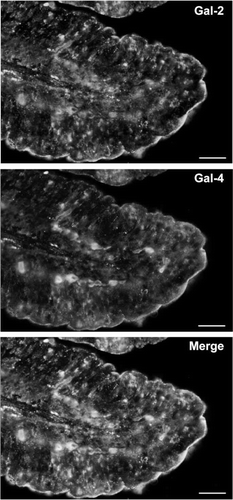
Figure 5. Release of microvillar galectin-2 and galectin-4 by different carbohydrates. (A) 50 µl samples of a microvillar DRF, prepared as described in Methods, were resuspended in 25 mM HEPES-HCl, 150 mM NaCl, pH 7.1, and incubated for 30 min at room temperature in the presence of 0 mM, 10 mM, 25 mM, 50 mM or 100 mM lactose, or 0 M, 0.25 M, 0.50 M, 1.0 M or 1.5 M of sucrose, fructose or maltose, respectively. After incubation, the samples were centrifuged at 20,000 g, 30 min, and the supernatant and pellet fractions subjected to SDS/PAGE and immunoblotting for galectins 2-and 4. (B) Samples of a right-side-out microvillar membrane vesicle preparation were resuspended in 25 mM HEPES-HCl, 150 mM NaCl, pH 7.1, in the absence of disaccharides (Con) or in the presence of 50 mM lactose (Lac), 0.5 M sucrose (Suc) or 0.5 M maltose (Mal). After incubation for 30 min at room temperature, the samples were centrifuged at 20,000 g for 30 min, and the supernatants collected and subjected to SDS/PAGE, followed by immunoblotting with an antibody to galectin-2.
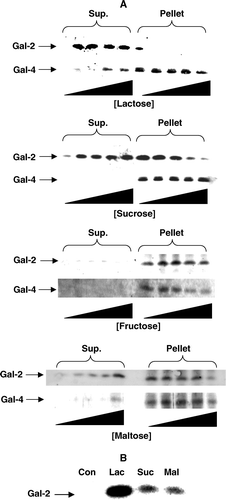
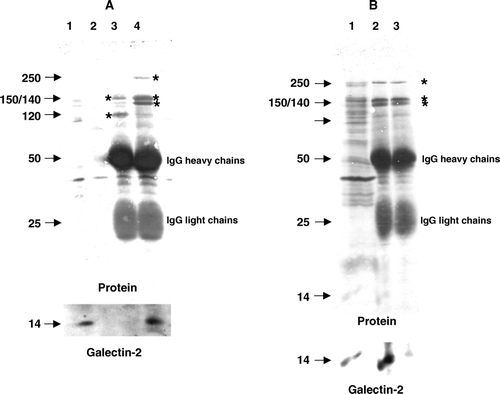
Figure 6. Coimmunopurification of galectin-2 with sucrase-isomaltase. (A) Aminopeptidase N and sucrase-isomaltase were immunoisolated from Triton X-100-solubilized DRF as described in Methods. After SDS/PAGE, galectin-2 was visualized by immunoblotting before total protein was stained with Coomassie Brilliant Blue. Lane 1; solubilized DRF, lane 2; protein A-Sepharose (control), lane 3; aminopeptidase N-coupled protein A-Sepharose, lane 4; sucrase-isomaltase-coupled protein A-Sepharose. The 150 and 120 kDa subunits of aminopeptidase N are seen in lane 3, and the 250 kDa single-chain sucrase-isomaltase, 150 kDa isomaltase and 140 kDa sucrase are seen in lane 4 (all marked by asterisks). (B) DRFs were solubilized by Triton X-100 as described in Methods and incubated for 30 min at room temperature in the absence or presence of 50 mM lactose before immunoisolation of sucrase-isomaltase. After SDS/PAGE, immunoblotting for galectin-2 was performed before staining for protein. (Asterisks mark the bands of sucrase-isomaltase.) Lane 1; solubilized DRF, lane 2; sucrase-isomaltase immunoisolated in the absence of lactose, lane 3; sucrase-isomaltase immunoisolated in the presence of lactose.