Figures & data
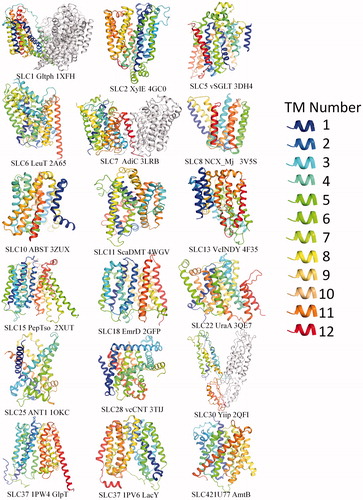
Table 1. Solute carrier family and the corresponding folds: solute carrier family and the corresponding function, folds and human diseases.
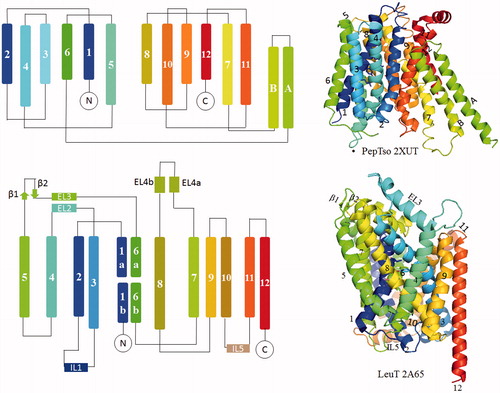
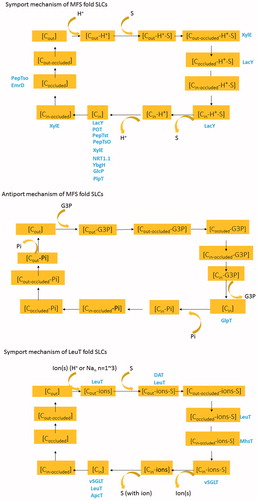
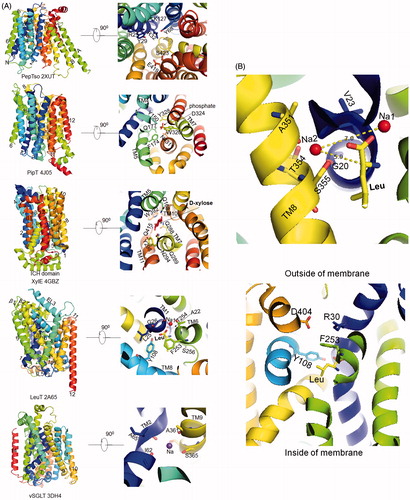
Table 2. Solute carrier family and the corresponding folds: categories of SLC structures based on folds.