Figures & data
Figure 1. (A) Plot of helix C of WT bacteriorhodopsin. The two proton transporting residues (Asp85 and Asp96) are indicated, as well as the location of the Schiff base. The steric clash between the ring Cγ of Pro91 and the carbonyl O of Leu87 is evidenced by using van der Waals spheres. (B) Cytoplasmic-to-extracellular view of helix C of bacteriorhodopsin. (C) Location of transmembrane prolines in bacteriorhodopsin. Van der Wals radii of the atoms involved in the steric clash are displayed. Cγ of proline residues are in white and the peptydil O of the i-4 residue is in red (PDB code 1PY6).
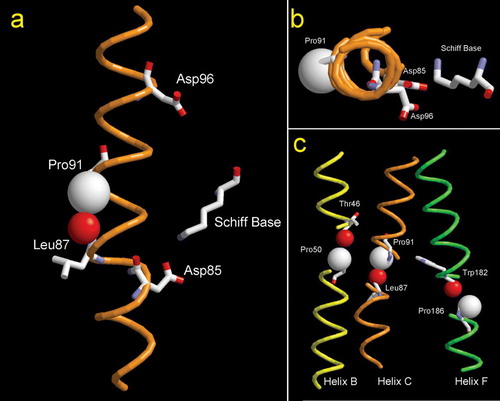
Table I. Spectral features of bacteriorhodopsin WT and proline mutants.
Figure 2. Hydroxylamine reactions. (Top) Rates of Schiff-base reaction with hydroxylamine in the absence of light recorded for WT (•) and mutants P50A (▪), P91A (▴) and P186A (★). (Bottam) Rates of Schiff-base reaction with hydroxylamine performed under illumination for WT (○), P50A (□), P91A (▵) and P186A (☆), using light of 300 lux of luminance. Plots correspond to changes of the intensity of the visible absorption band as a function of time.
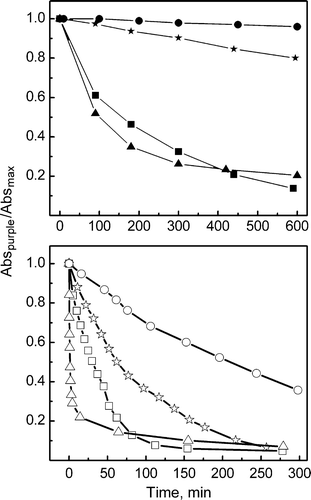
Figure 3. Kinetics of M formation and decay. Changes in absorbance at 410 nm of WT, P50A, P91A and P186A suspensions in 1M KCl, pH 6.5 at room temperature after a ns laser flash were averaged and plotted as a function of time. The time constants of M rise (τave) were: WT 60 µs, P50A 45 µs, P91A 7 µs and P186A 35 µs.
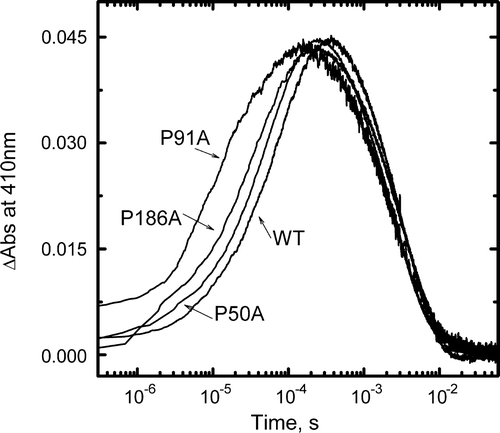
Figure 4. Light-induced pH changes as measured with the pH-sensitive dye pyranine. Pyranine absorption changes were measured at 460 nm in suspensions of WT, P50A, P91A and P186A in 1M KCl (pH 7.2) at 298 K as a function of time, after laser flash excitation. Proton uptake from BR causes an increase of absorbance, while proton release causes a decrease. Protein concentration, 1.5×10−5 M.
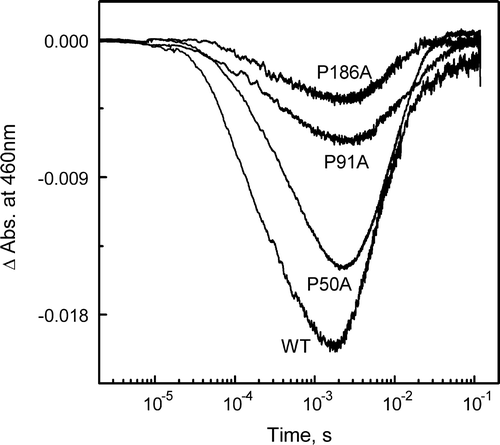
Table II. Bacteriorhodopsin relative proton pumping activity.
Figure 5. Light-induced pH changes of BR-incorporated liposomes as a function of time. The samples correspond to WT (•), P50A (▪), P91A (▴) and P186A (★). Initial pH 6.5, in KCl 150 mM.
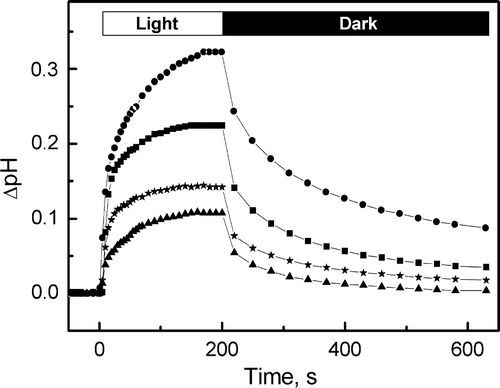
Figure 6. Fourier transform infrared difference spectra corresponding to M intermediate state minus the BR resting state. Spectra for WT, P50A, P91A and P186A were collected under M-yielding conditions (150 mM KCl, dry film, pH 10, 243 K). Spectra shown are the addition of 1050 interferograms taken at a resolution of 2 cm−1.
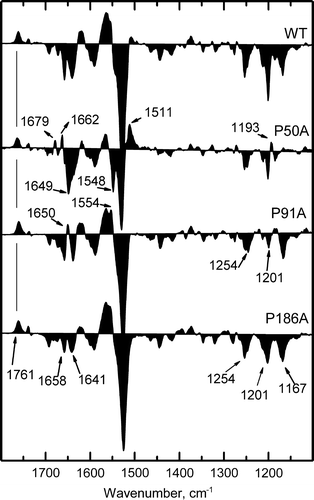
Figure 7. Fourier transform infrared difference spectra corresponding to N intermediate state minus the BR resting state. Spectra for WT, P50A, P91A and P186A were collected under N-yielding conditions (150 mM KCl, wet film, pH 10, 277 K). Spectra shown are the addition of 1050 interferograms taken at a resolution of 2 cm−1.
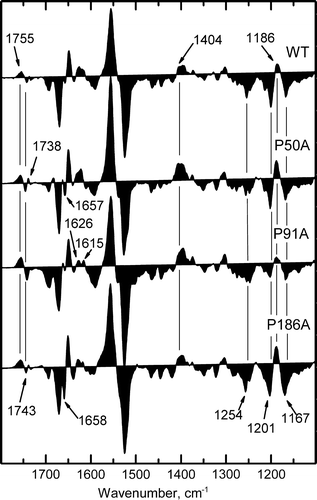
Table III. Distances in Å between Cδi of Pro (50, 91 and 186) and the peptidyl O of the i-3 and i-4 residues of bacteriorhodopsin.