Figures & data
Figure 1. Biosynthesis of glycosylphosphatidylinositol (GPI)-glycoconjugates. Shown is the GPI biosynthesis pathway in eukaryotes. Steps that are unique to the respective organism are indicated as follows: yyeast; mmammalian cells (reviewed in Citation[134]). GIPLs and the GPI-phosphosaccharides, e.g., lipophosphoglycan (LPG) or lipophosphopeptidoglycan (LPPG), are formed from distinct intermediates in the GPI biosynthesis pathway, designated, respectively as PI2 and M1 (Manα1-4GlcN-PI) Citation[135]. Specific enzymatic reactions: (1) GlcNAc transferase complex, addition of GlcNAc to PI; (2) GlcNAc-PI-de-N-acetylase, removal of N-acetyl group from GlcNAc; (2A) inositol-acyltransferase, addition of acyl group to GlcN-PI at position 2y,m; (3) GPI-α1-4mannosyltransferase (GPI-α1-4MT), transfer of mannose to position 4 of GlcN; (F) Flippase, translocation of GPI intermediate from the cytoplasmic to the luminal face of endoplasmic reticulum; (3A) Ethanolamine phosphotransferase (EtNPT-I), addition of EtNP to the first mannosey,m; (4) GPI-α1-6MT, transfer of mannose to position 6 of the first mannose; (5) and (5A) GPI-α1-2MT, transfer of mannose to position 2 of the second and thirdy mannose, respectively; (6) EtNPT-III, transfer of EtNP to the third mannose at position 6; (6A) EtNPT-II, transfer of EtNP to the second mannose at position 6y; (7) Transamidase complex, cleavage of C-terminal GPI addition signal sequence (ss) of the precursor protein and attachment of GPI. This Figure is reproduced in color in Molecular Membrane Biology online.
![Figure 1. Biosynthesis of glycosylphosphatidylinositol (GPI)-glycoconjugates. Shown is the GPI biosynthesis pathway in eukaryotes. Steps that are unique to the respective organism are indicated as follows: yyeast; mmammalian cells (reviewed in Citation[134]). GIPLs and the GPI-phosphosaccharides, e.g., lipophosphoglycan (LPG) or lipophosphopeptidoglycan (LPPG), are formed from distinct intermediates in the GPI biosynthesis pathway, designated, respectively as PI2 and M1 (Manα1-4GlcN-PI) Citation[135]. Specific enzymatic reactions: (1) GlcNAc transferase complex, addition of GlcNAc to PI; (2) GlcNAc-PI-de-N-acetylase, removal of N-acetyl group from GlcNAc; (2A) inositol-acyltransferase, addition of acyl group to GlcN-PI at position 2y,m; (3) GPI-α1-4mannosyltransferase (GPI-α1-4MT), transfer of mannose to position 4 of GlcN; (F) Flippase, translocation of GPI intermediate from the cytoplasmic to the luminal face of endoplasmic reticulum; (3A) Ethanolamine phosphotransferase (EtNPT-I), addition of EtNP to the first mannosey,m; (4) GPI-α1-6MT, transfer of mannose to position 6 of the first mannose; (5) and (5A) GPI-α1-2MT, transfer of mannose to position 2 of the second and thirdy mannose, respectively; (6) EtNPT-III, transfer of EtNP to the third mannose at position 6; (6A) EtNPT-II, transfer of EtNP to the second mannose at position 6y; (7) Transamidase complex, cleavage of C-terminal GPI addition signal sequence (ss) of the precursor protein and attachment of GPI. This Figure is reproduced in color in Molecular Membrane Biology online.](/cms/asset/bdc60b6e-b242-45e3-a6e2-e3ac9d34fa10/imbc_a_160132_f0001_b.jpg)
Figure 2. Characteristic features of the proteins anchored by GPI. Precursor proteins destined to receive a GPI contain two signal sequences, an N-terminal signal sequence and a C-terminal signal sequence, which respectively direct the translocation of the protein to the endoplasmic reticulum (ER) and the anchoring of the GPI to the protein. The C-terminal signal sequence is composed of cleavage attachment (C/A) site of three amino acids, referred to as ω (omega), ω + 1, ω + 2; a short hydrophilic spacer element (designated ---); and a series of hydrophobic amino acids (hydrophobic domain). Cleavage occurs between the ω and ω + 1 site and is followed by GPI attachment Citation[76].
![Figure 2. Characteristic features of the proteins anchored by GPI. Precursor proteins destined to receive a GPI contain two signal sequences, an N-terminal signal sequence and a C-terminal signal sequence, which respectively direct the translocation of the protein to the endoplasmic reticulum (ER) and the anchoring of the GPI to the protein. The C-terminal signal sequence is composed of cleavage attachment (C/A) site of three amino acids, referred to as ω (omega), ω + 1, ω + 2; a short hydrophilic spacer element (designated ---); and a series of hydrophobic amino acids (hydrophobic domain). Cleavage occurs between the ω and ω + 1 site and is followed by GPI attachment Citation[76].](/cms/asset/15b0d4b9-0b4f-4ba9-87d2-b1de9cd2d029/imbc_a_160132_f0002_b.gif)
Table I. Characteristics of GPI8s.
Figure 3. The postulated catalytic mechanism for GPI-anchor attachment by GPI8. Cleavage of the C-terminal signal sequence of the precursor protein is believed to occur in an endoproteolytic reaction in which the peptide bond is cleaved between the ω and ω + 1 site within the protein chain, followed by GPI attachment to the protein in an amidation reaction. Cysteine proteases (enzyme, Enz) perform peptide bond hydrolysis via a general acid: base catalysis, as follows: (1) The sulfhydryl (SH) group of the cysteine (Cys) residue is deprotonated by the histidine (His) residue, which acts as a ‘general base’ or proton acceptor; (2) Nucleophilic attack of the Cys's sulfur on the carbonyl carbon results in the cleavage of the peptide and the formation of an acyl-enzyme intermediate. The transfer of acyl to water (H2O) that serves as a nucleophile releases the Enz and formation of carboxylic acid on the cleaved peptide. When GPI serves as the nucleophile, an amide bond (*) is formed between the nitrogen in the ethanolamine phosphate (EtNP) of the GPI and the carbonyl group of the protein, completing the attachment of a GPI to the protein. Alkyl groups of the protein are represented as R; R1 corresponds to the portion of the precursor protein that is GPI-anchored; R2 corresponds to the peptide or, in the case of protein-GPI anchoring, the C-terminal signal sequence that is released. (Diagram created in ChemSketch 5.0, Advanced Chemistry Development Inc.).
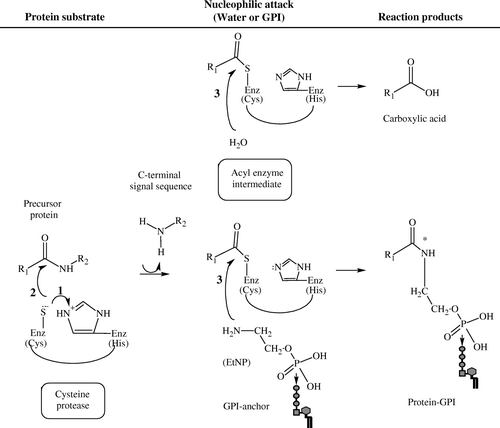
Figure 4. CLUSTAL W (1.82) multiple sequence alignment of GPI8 in T. cruzi, T. brucei, L. mexicana, S. cerevisiae, H. sapiens, T. gondii and P. falciparum and of legumain in Canavalia ensiformis. Accession numbers: TbGPI8, AJ308106; LmGPI8, AJ242865; yGPI8, P49018; hGPI8, Q92643; TgGPI8, AJ507036; PfGPI8, AJ401201; Legumain, JX0344. Arrows indicate the conserved cysteine (C) and histidine (H) residues. The postulated active site residues for GPI8, based on mutagenesis studies of legumain (plant endopeptidase), are indicated by •. Specific residues that have been examined by mutagenesis and overexpression in the respective organisms are underlined.
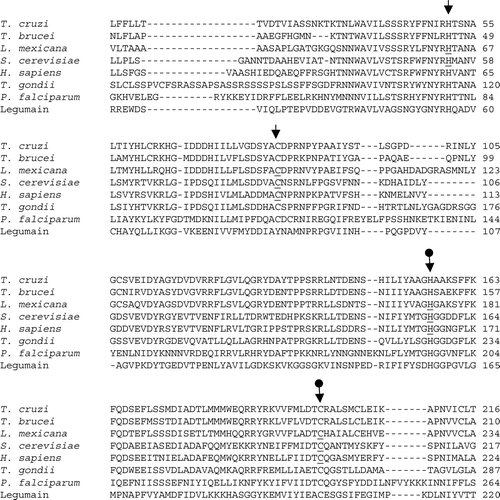
Figure 5. The transamidase (TAM) complex. A model of GPI8 interaction with other proteins in the TAM complex is presented for (A) yeast and human cells, and (B) T. brucei. Structures are not drawn to scale. Identification and functional analysis of the indicated proteins is described in the text Citation[124]. Adapted by permission from Macmillan Publishers Ltd: EMBO Journal 20(15):4088–4098, copyright 2001.
![Figure 5. The transamidase (TAM) complex. A model of GPI8 interaction with other proteins in the TAM complex is presented for (A) yeast and human cells, and (B) T. brucei. Structures are not drawn to scale. Identification and functional analysis of the indicated proteins is described in the text Citation[124]. Adapted by permission from Macmillan Publishers Ltd: EMBO Journal 20(15):4088–4098, copyright 2001.](/cms/asset/53d4edaa-989c-4a91-9723-09da78e0ccdf/imbc_a_160132_f0005_b.gif)
Table II. Other components of GPI-anchoring machinery.