Figures & data
Figure 1. Arabidopsis contains two PEX19 genes. Alignment of Arabidopsis PEX19-1 and 19-2 with human PEX19 and Saccharomyces cerevisiae PEX19. Amino acid sequences were aligned with the programme T-coffee Citation[43]. Grey shading indicates hydrophobic residues in conserved regions that are predicted to form amphipathic helices. The CAAX motif directing farnesylation is underlined and cysteine residues are indicated in bold.
![Figure 1. Arabidopsis contains two PEX19 genes. Alignment of Arabidopsis PEX19-1 and 19-2 with human PEX19 and Saccharomyces cerevisiae PEX19. Amino acid sequences were aligned with the programme T-coffee Citation[43]. Grey shading indicates hydrophobic residues in conserved regions that are predicted to form amphipathic helices. The CAAX motif directing farnesylation is underlined and cysteine residues are indicated in bold.](/cms/asset/cb793e71-a5cf-47f1-bf67-ba41f77e8165/imbc_a_173785_f0001_b.gif)
Figure 2. Expression profiles of Arabidopsis PEX19-1 and PEX19-2 in different tissues. Starting quantity values as determined from a standard curve derived from a serial dilution of a pool of cDNA comprising equal volumes of each cDNA are plotted for each tissue. Each value is the mean±SE of six values (2 biological replicates with 3 technical replicates of each sample), except PEX19-1 in silique which is the mean±SE of 5 values (2 biological replicates with 2 and 3 technical replicates respectively). DG, LG stands for dark grown and light grown seedlings respectively; CC stands for cell culture.
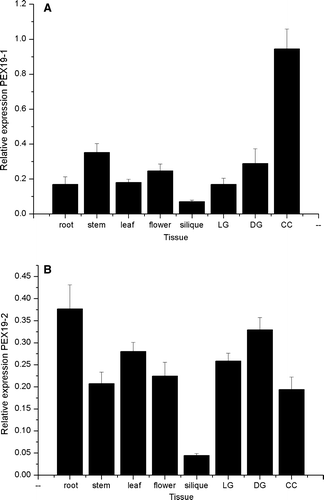
Figure 3. Arabidopsis PEX19 is predominantly a cytosolic protein. (A) Anti-PEX19 antibodies recognize native PEX19, a 30 kDa protein in Arabidopsis seedlings. Left panel, immunoblot probed with anti-PEX19 anti-serum 1:10,000 dilution; lane 1, 10 ng recombinant His6T7PEX19, lane 2, 1 ng His6T7PEX19, lane 3, total protein extract equivalent to 20 2-day-old Arabidopsis seedlings. Right panel, as for left panel except that the anti-PEX19 antiserum was preincubated with 10 µg His6T7PEX19 prior to immunodecoration of the blot. (B) PEX19 is mainly cytosolic. Lysate of tissue culture cells (T) was separated into a soluble fraction (259,000 g supernatant; S), and a membrane associated fraction (259,000 g pellet; M). Equivalent volumes were separated by SDS-PAGE and probed with anti-PEX19 serum. (C) A post nuclear supernatant prepared from tissue culture cells was separated by sucrose density gradient centrifugation and equal volumes of each fraction blotted and detected with anti-PEX19 (top panel) and anti-thiolase (lower panel). The graph shows protein concentration (mg/ml) and sucrose concentration (% w/v) in the various fractions.
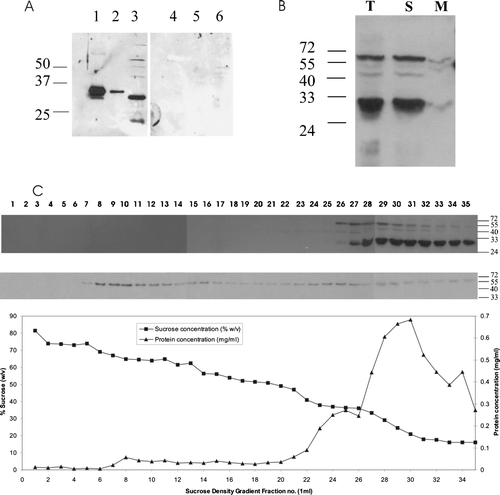
Figure 4. Recombinant His6PEX19-1 forms a dimer. (A) His6PEX19-1 that had been purified by nickel affinity chromatography was applied to a Superdex 75 20/60 gel filtration column and eluted with a retention volume of 115 ml (shown by downward pointing arrow) indicating it has a mass of 60 kDa, consistent with being a dimer. The column was calibrated using a gel filtration low molecular weight calibration kit (GE Health Care) with retention volumes and masses of the protein standards indicated on the chromatogram (upward pointing arrows). For both standards and sample, the retention volume where each protein begins to elute from the column is indicated. V0 is the void volume. (B) Coomassie stained SDS-PAGE showing His6PEX19-1 in fractions eluted from the gel filtration column. His6PEX19-1 in fractions collected between 110 and 134 ml (indicated by broken double arrows on A) show that it migrates with an apparent mass of approximately 30 kDa in SDS-PAGE, despite elution from the column with a mass of approximately 60 kDa. The lane indicated as P shows nickel affinity column purified His6PEX19 before gel filtration chromatography. (C) Immunoblot of 12% native Tris/glycine gels, showing equivalent volumes of total (T), soluble (S) and membrane (M) fractions, (see B), extracted in the presence (+) or absence (-) of DTT and probed with anti-AtPEX19 antibody and (D) anti-AtPEX19 antibody, preincubated with recombinant PEX19 as in B.
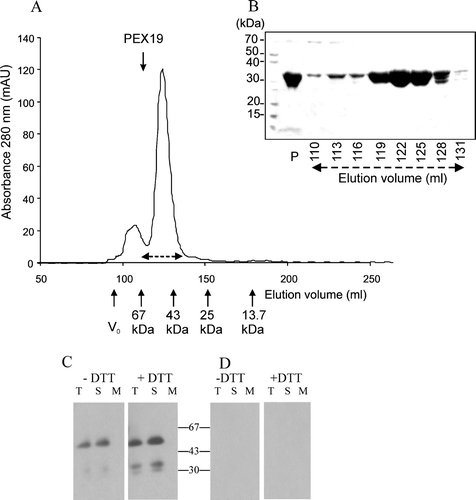
Figure 5. In vitro translated AtPEX19-1 interacts with PEX10. His6T7PEX19-1 and PEX10 were in vitro translated and mixed (lane B) prior to immunoprecipitation by non-immune serum (PI) or anti-His antibodies (H) followed by SDS-PAGE and phosphoimaging. Solid arrow indicates the position of His6T7PEX19-1; open arrow indicates the position of PEX10. 14C Molecular weight standards are, 97 kDa (doublet), 66 kDa, 45 kDa, 30 kDa, 20.1 kDa..
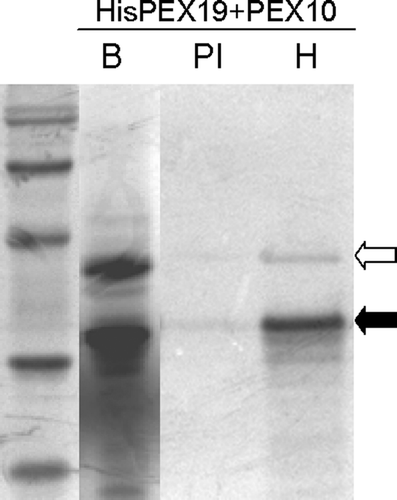
Figure 6. Recombinant His6PEX19-1 binds to the C-terminus of PEX10. Pull down experiments using lysates from E. coli expressing His6PEX19-1 (PEX19) or GST-PEX10267-381 (PEX10) were performed; The lysates were applied alone or as a mixture of both, to a glutathione column, the flow through collected, column washed and specifically bound material was eluted with 10 mM reduced glutathione. The fractions (P, sample prior to column application, FT, flow through, W, wash and E, eluate) were analysed by Western blot, immuno-stained with anti-PEX19 (panel A) or anti-GST (panel B) antibodies. Positions of His6PEX19-1 (panel A) and GST-PEX10267-381 (panel B) are indicated by arrows
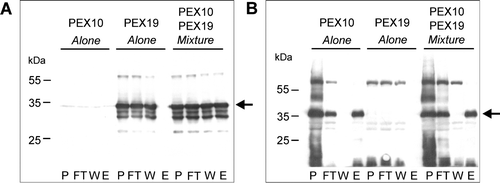
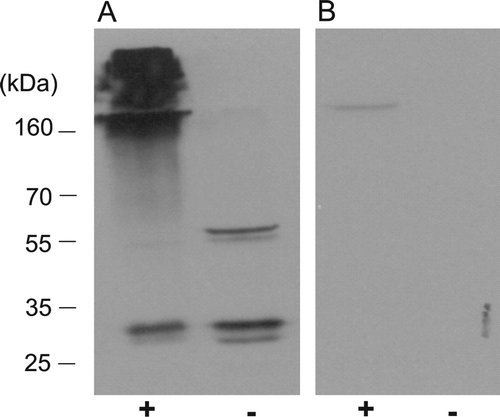
Figure 7. The dimeric form of PEX19 is preferentially cross-linked to other proteins in vivo. Arabidopsis suspension cells were cross-linked in vivo with 1% formaldehyde. (A) The cell lysates, cross-linked (lane +), or not cross-linked (lane -), were examined by immunoblotting using antiserum raised against recombinant His6PEX19-1 at 1:10,000 dilution. (B) As panel A, but the antisera was incubated with rHis6PEX19 (10? µg) for 30 min at 37°C before immunodecoration of the blot.
Supplementary Figure 1. Expression profiles of Arabidopsis PEX19-1. Expression profiles were constructed from public microarray data using tools available on the GENEVESTIGATOR website Citation[44]. (A) Expression in various plant organs generated by the Gene Atlas tool. (B) Expression at different developmental stages, defined by Citation[45]. Stage 0, seed germination; stage 1, leaf development; stage 3, rosette growth; stage 5, inflorescence emergence; stage 6, flower production; stage 8, silique development generated by the gene chronologer tool.
![Supplementary Figure 1. Expression profiles of Arabidopsis PEX19-1. Expression profiles were constructed from public microarray data using tools available on the GENEVESTIGATOR website Citation[44]. (A) Expression in various plant organs generated by the Gene Atlas tool. (B) Expression at different developmental stages, defined by Citation[45]. Stage 0, seed germination; stage 1, leaf development; stage 3, rosette growth; stage 5, inflorescence emergence; stage 6, flower production; stage 8, silique development generated by the gene chronologer tool.](/cms/asset/add4f68d-ea47-451c-9517-1a40be0a4739/imbc_a_173785_f0008_b.gif)