Figures & data
Figure 1. Adjacent subunits (principal and complementary) showing the positions of Asn-128, Glu-129 and Phe-130 in the original homology model of the 5-HT3 receptor (Reeves et al. [Citation2003], Thompson et al. [Citation2005]). (A) Only two of the five subunits have been shown for ease of viewing. In the right hand panel the complementary subunit has been removed and the binding site rotated to view it from the side. (B) An alignment of the 5-HT3A, nAChR α1 and AChBP receptor subunit sequences. The binding loops of the receptors are indicated by black lines above the alignment and their location can be seen in the structure above. The positions of the β-sheets are shown by grey lines beneath the text and are taken from the AChBP protein crystal structure (Brejc et al. [Citation2001]). Asn-128, Glu-129 and Phe-130 are highlighted as white text in a black box. Accession numbers for the 5-HT3A, nACh α1 and AChBP subunit sequences are Q6J1J7, P02710 and P58154 respectively.
![Figure 1. Adjacent subunits (principal and complementary) showing the positions of Asn-128, Glu-129 and Phe-130 in the original homology model of the 5-HT3 receptor (Reeves et al. [Citation2003], Thompson et al. [Citation2005]). (A) Only two of the five subunits have been shown for ease of viewing. In the right hand panel the complementary subunit has been removed and the binding site rotated to view it from the side. (B) An alignment of the 5-HT3A, nAChR α1 and AChBP receptor subunit sequences. The binding loops of the receptors are indicated by black lines above the alignment and their location can be seen in the structure above. The positions of the β-sheets are shown by grey lines beneath the text and are taken from the AChBP protein crystal structure (Brejc et al. [Citation2001]). Asn-128, Glu-129 and Phe-130 are highlighted as white text in a black box. Accession numbers for the 5-HT3A, nACh α1 and AChBP subunit sequences are Q6J1J7, P02710 and P58154 respectively.](/cms/asset/4984cf30-da45-4a9c-a7a7-0385d1838a44/imbc_a_183090_f0001_b.gif)
Table I. [3H]-Granisetron binding affinities.
Figure 2. Non-binding Glu-129 mutant 5-HT3 receptors expressed in HEK 293 cells. Levels of expression in permeabilized cells (left hand column) were similar to wild type for all mutants (20–40% of cells were fluorescently labelled), but only reached these levels in non–permeabilized cells (right hand column) for E129G and E129A receptors. Fluorescent cell surface labelling was present in < 2% cells for all other mutants, although, as shown, some receptors did appear to reach the plasma membrane in some cells. Scale bars indicate 10 µm.
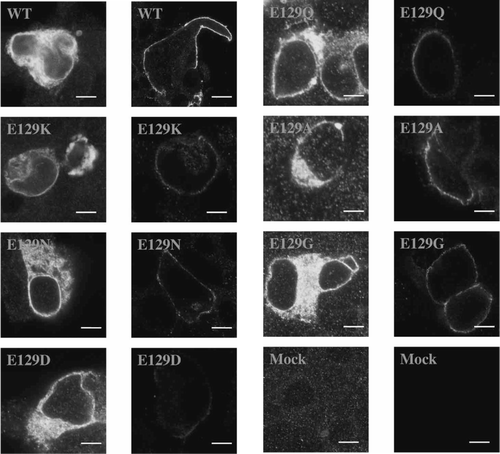
Figure 3. Dose-response curves derived from FlexStation responses to 5-HT stimulation. Data have been normalized to the maximum fluorescence absorbance (Fmax) and plotted as the mean±SEM. (A) Asn-128 mutants. (B) Phe-130 mutants. Inset: Typical Flexstation responses to 5-HT (0, 0.1, 0.5 or 1.0 µM). 5-HT was added at 20 sec to HEK 293 cells expressing F130Y-5-HT3 receptors. F = fluorescence in arbitrary units; t=time in seconds.
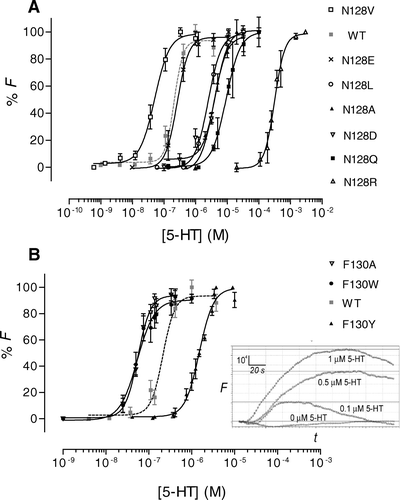
Table II. Functional parameters.
Figure 4. Original and new models of the 5-HT3 receptor extracellular domain with docked 5-HT and granisetron. (A) Subunit dimer showing the ligand binding site (circled) at the subunit interface. (B) Close-up of the ligand binding site of the original model showing residues Asn-128, Glu-129 and Phe-130 of binding loop A in relation to docked 5-HT (from Reeves et al. [Citation2003]) and granisetron (from Thompson et al. [Citation2005]). (C) New model 1 with residues moved closer to the N-terminus so that Glu-129 now points towards the binding site. (D) New model 2 with residues moved further towards the N-terminus so that Phe-130 now points towards the binding site.
![Figure 4. Original and new models of the 5-HT3 receptor extracellular domain with docked 5-HT and granisetron. (A) Subunit dimer showing the ligand binding site (circled) at the subunit interface. (B) Close-up of the ligand binding site of the original model showing residues Asn-128, Glu-129 and Phe-130 of binding loop A in relation to docked 5-HT (from Reeves et al. [Citation2003]) and granisetron (from Thompson et al. [Citation2005]). (C) New model 1 with residues moved closer to the N-terminus so that Glu-129 now points towards the binding site. (D) New model 2 with residues moved further towards the N-terminus so that Phe-130 now points towards the binding site.](/cms/asset/ad135f41-72b8-4adc-a900-e5117f7bf875/imbc_a_183090_f0004_b.jpg)