Figures & data
Figure 1. Bilayer reconstitution of CLIC4 in the presence of 1 mM DTT. Part A shows examples of recordings at selected holding potentials, with solid lines to indicate the closed levels, and an example of an all-points amplitude histogram from 30 sec of the +100 mV recording (the arrow indicates the maximum open level). Part B summarizes the I/V relationship of the main open level and a ∼25% substate, from 15 independent experiments with 500 mM KCl cis vs. 50 mM KCl trans. The bars indicate ±1 SD, and the line was fitted by eye (see text for linear regression analysis). The inset shows examples of direct transitions between the ∼25% substate, the main open level and the closed level, on an expanded time scale.
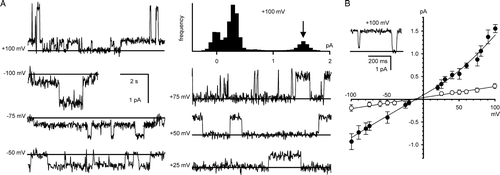
Figure 2. Conductance/activity relationship of CLIC4. The data in the main panel (shown as means±SD from 5 independent experiments) were fitted (by least squares) to the sum of a hyperbolic component with a gmax of 8.2 pS and a “Km” of 19.1 mM plus a non-saturating linear component of 0.017 pS per mM KCl. The relationship breaks down at KCl activities >325 mM (as shown by the dotted lines – note the break in the plot between 400–600 mM KCl). The inset I/V relationships provide examples of the I/V data used to calculate the conductance at low (40 mM, closed circles) and high (325 mM, open circles) activities (points are means±SD, n=5, lines fitted by linear regression).
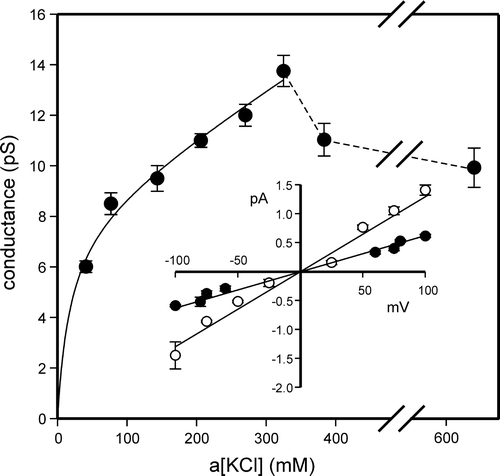
Figure 3. CLIC4 currents in Tris-HCl (pH 7.4). Examples of CLIC4 currents (part A) and a combined I/V relationship from several independent experiments (part B, means±1 SD, n=7) in the presence of 1 mM DTT with 500 mM TrisCl cis vs. 50 mM TrisCl trans. The line was fitted by linear regression.
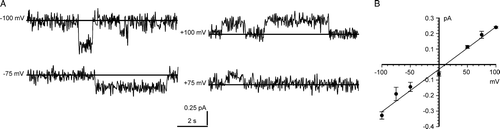
Figure 4. Bilayer reconstitution of CLIC4 in the presence of 100 µM H2O2. Part A shows examples of recordings at selected holding potentials, and an example of an all-points amplitude histogram from 30 sec of the +100 mV recording (the arrow indicates the maximum open level). Part B summarizes the I/V relationship obtained from 5 independent experiments (means±1 SD, n=5). The line was fitted by eye (see text for linear regression analysis). All the data were obtained with 500 mM KCl cis vs. 50 mM KCl trans.
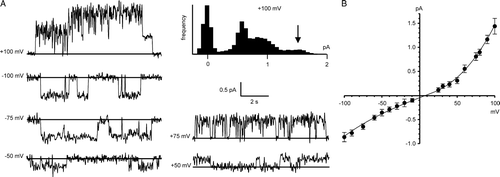
Figure 5. Bilayer reconstitution of CLIC4 in glutathione buffers. Part A shows recordings in 5 mM GSH (cis and trans), with GSSG added sequentially to the trans chamber to give the indicated GSH/GSSG ratios. Part B shows a combined I/V relationship from 5 independent experiments (means±SD) with 5 mM GSH/0.5 mm GSSG both cis and trans The line was fitted by eye (see text for results of linear regression analysis). The traces in part C (underfiltered, and on an expanded time scale), illustrate the appearance of substates (best-resolved where indicated by the arrowhead) immediately after replacing the cis glutathione buffer (5 mM GSH/0.5 mM GSSG) with 1 mM DTT by perfusion. All the data were obtained with 500 mM KCl cis vs. 50 mM KCl trans, and the recordings were carried out at a HP of +100 mV.
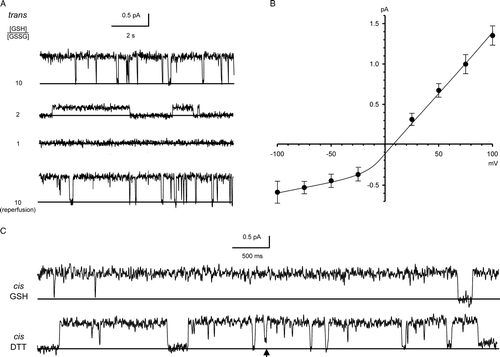
Figure 6. FPLC and SDS-PAGE analysis of full-length and truncated CLIC4. The main panel shows typical gel-exclusion FPLC profiles for full-length (FL) and truncated (TR) proteins in the presence of 5 mM DTT (dashed and solid lines, respectively, shown on the same plot for comparison). Fractions from the boxed peaks (I and II, respectively) were collected and concentrated, and subjected to reducing 10% (w/v) SDS-PAGE and Coomassie-stained as shown. FL and TR proteins show Mr values of ∼30 K and ∼6–7 K, respectively. The FPLC analysis reveals similar values (Vt was 70 ml, Vo was 4.5 ml, and the Kav values for the FL and TR proteins are 0.73 and 0.47, respectively). The column was calibrated (inset plot) with: RNase A (13.7 K), pGEX vector GST (26 K), ovalbumin (43 K) and albumin (67 K).
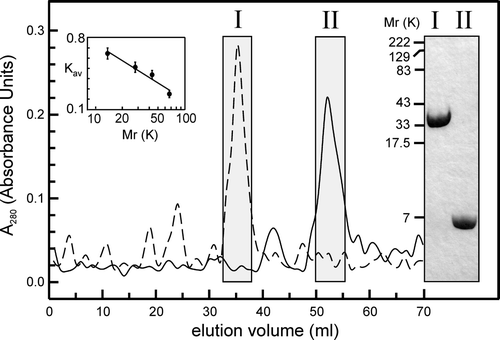
Figure 7. Bilayer reconstitution of truncated CLIC4. Part A compares the appearance of channels at +100 mV in the presence of either 1 mM DTT, 100 µM H2O2 (with a star to indicate an unusual large-amplitude opening, see text), or the indicated GSH/GSSG ratios (with [GSH] set to 5 mM), all in a cis:trans gradient of 500:50 mM KCl. Part B summarizes the corresponding I/V relationships in the presence of 1 mM DTT (closed circles), 100 µM H2O2 (open circles) and a 10:1 GSH/GSSG buffer (open squares) (means±1 SD for 5 independent experiments in each case). The line was fitted by eye (see text for results of linear regression analysis).
![Figure 7. Bilayer reconstitution of truncated CLIC4. Part A compares the appearance of channels at +100 mV in the presence of either 1 mM DTT, 100 µM H2O2 (with a star to indicate an unusual large-amplitude opening, see text), or the indicated GSH/GSSG ratios (with [GSH] set to 5 mM), all in a cis:trans gradient of 500:50 mM KCl. Part B summarizes the corresponding I/V relationships in the presence of 1 mM DTT (closed circles), 100 µM H2O2 (open circles) and a 10:1 GSH/GSSG buffer (open squares) (means±1 SD for 5 independent experiments in each case). The line was fitted by eye (see text for results of linear regression analysis).](/cms/asset/5d4a412d-164d-40e7-84e8-cca5fa11ad38/imbc_a_192707_f0007_b.gif)
Figure 8. Inhibition of CLIC4 currents by DTNB. Parts A and B show examples of CLIC4 currents from full-length and truncated proteins, respectively, at +100 mV, obtained in the presence of 5 mM GSH (with 500 mM KCl cis vs. 50 mM KCl trans) before removing the GSH by perfusion with fresh GSH-free solutions. In each case, 0.2 mM DTNB had no effect from the cis side, but abolished channel activity when added from the trans side. See text for full statistical analysis.
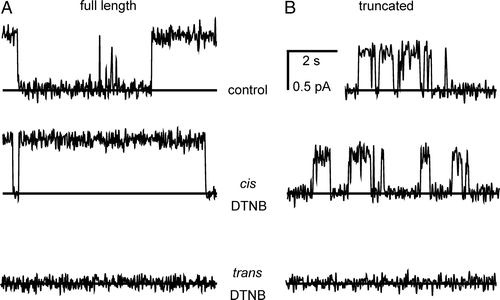
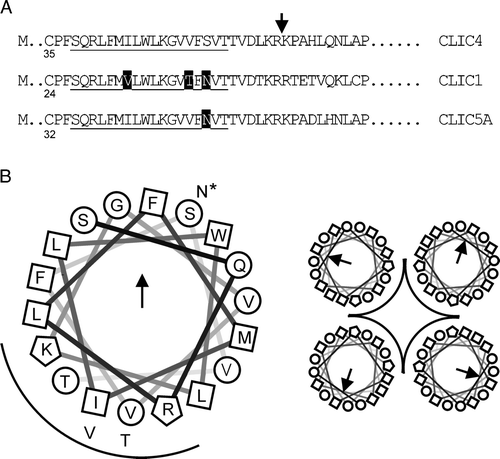
Figure 9. Organization of the putative CLIC TMD. Part A: selected regions of 3 CLICs aligned at the putative 18-residue TMD (underlined). The first cysteine residue in each sequence is numbered, and differences within the TMD are highlighted (rat and human CLIC4 are identical in this region). CLIC4 was truncated where indicated. Part B is a helical wheel projection of the TMD from N to C (D. Armstrong and R. Zidovetzki, http://rzlab.ucr.edu/scripts/wheel/wheel.cgi) with a “tetrameric pore” cartoon. Squares, circles and polygons indicate relatively hydrophobic or hydrophilic residues, or residues with positively charged side chains, respectively. The arrow and the arc indicate the direction of the hydrophobic moment, and suggested pore-lining residues, respectively. The alternative residues in CLIC1 (all 3) and CLIC5A (starred residue only) are also shown.