Figures & data
Figure 1. Opposite orientation of N and C termini in CLN6. BHK21 cells transiently expressing the C-terminally myc-tagged CLN6 fusion protein were fixed and membranes either permeabilized with Triton X-100 (a–c) or selectively with 1 µg/ml digitonin (d–f). The N terminus was detected using purified primary CLN6 antibody 1747, and secondary rhodamine-conjugated antibody (a and d). The C terminus was visualized with the anti-myc and a secondary FITC-conjugated antibody (b and e). The images were merged (c and f). Yellow indicates the simultaneous accessibility of primary antibodies to the cytosolic and luminal epitopes. Bars, 5 µm. This Figure is reproduced in colour in Molecular Membrane Biology online.
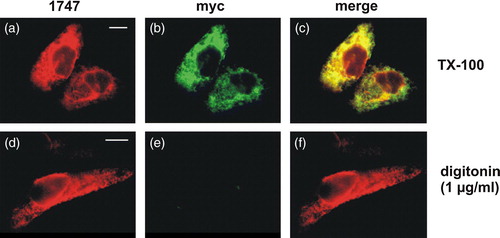
Figure 2. ER retention of CLN6 is not mediated by dibasic or dileucine-motifs. (A) Wild type (a) and various CLN6myc constructs (b–h) are represented schematically. The triple arginine motif in the N terminus was substituted by alanine residues (p.Arg5_Arg7Ala = b) or sequential N-terminal truncations were introduced (p.Glu2_Leu10del; p.Glu2_Leu20del; p.Glu2_Phe49del = c–e). Potential retention motifs in the C terminus of CLN6 were substituted by alanine residues (p.Lys288_Lys289Arg; p.Leu305_His306Ala g, h). (B) The wild type (a) and mutant CLN6myc constructs (b–h) were transiently expressed in BHK21 cells, stained for the myc-tag in red, and double-labeled with an antibody against PDI (green). Merged images (yellow) indicate overlapping localization of CLN6myc constructs with the ER marker. Bars, 5 µm. Figure 2B is reproduced in colour in Molecular Membrane Biology online.
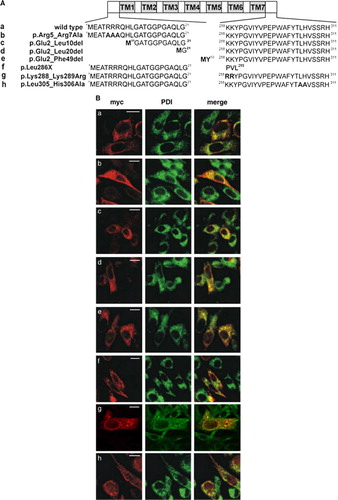
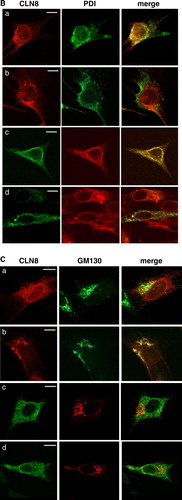
Figure 3. The N terminus of CLN6 mediates ER retention. (A) Schematic representation of CLN constructs used in the experimental approach: the ER-resident wild type (a) and the mutant CLN8 (K283R/K284R = b) protein localized in the Golgi apparatus, and the respective CLN6-CLN8 fusion proteins in which the N terminally 24 amino acids of CLN8 were substituted by the 49 amino acid N terminus of CLN6 (c, d). The position of the dibasic motif (KK) in the C-terminal domain of CLN8 is indicated. (B and C) the CLN8 (a, b) and CLN6-CLN8 constructs (c, d) were expressed in BHK21 cells and visualized using a polyclonal antibody directed against the C terminus (amino acid residues 268–286) of CLN8 (red). The cells were double-labeled with monoclonal antibodies against PDI (B) or GM130 (C) shown in green. Yellow indicates in the merged images overlapping of CLN constructs with the ER or Golgi markers. Bars, 5 µm. B and C are reproduced in colour in Molecular Membrane Biology online.
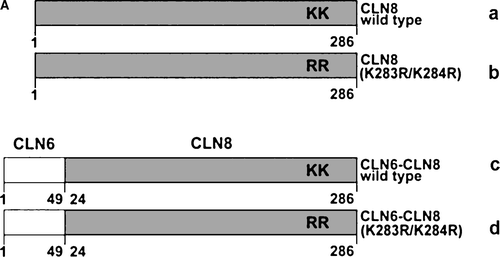
Figure 4. Expression of N- and C-terminal truncated CLN6 mutants. (A) Schematic representation of CLN6myc truncation mutants affecting the N terminus (p.Glu2_Leu20del = 2; p.Glu2_Phe49del = 3; p.Glu2_Ile118del = 4) or the C terminus (p.Leu286X = 5; p.Glu225X = 6). (B) The expression of the wild type and CLN6myc mutants were analysed by Western blotting using anti-myc antibodies 24 h after transfection of BHK21 cells.
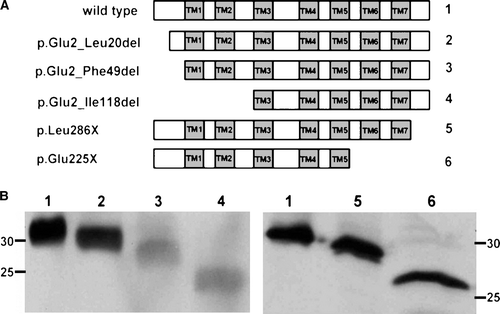
Figure 5. The distal pair of transmembrane domains of CLN6 represent a dominant ER retention structure. The localization of N- and C-terminally truncated CLN6myc mutants (p.Glu2_Ile118del and p.Glu225X corresponding to construct number 4 and 6, respectively, described in A) were analysed by double immunofluorescence microscopy 24 h after transfection using either the anti-myc antibody or antibodies against PDI, or GM130. Blue staining represents nuclear 4’, 6 diamidino-2-phenylindole (DAPI) staining used to show location of the cells. Bars, 10 µm. This Figure is reproduced in colour in Molecular Membrane Biology online.
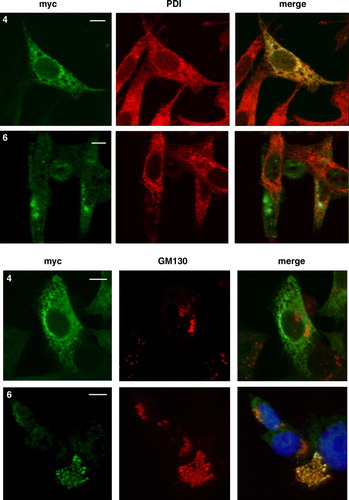
Figure 6. Localization of wild type and mutant CLN6 forms in hippocampal neurons. Mouse hippocampal primary neurons were transfected with CLN6 cDNAs encoding wild type (1) or p.Glu2_Phe49del (3) and p.Glu225X (6) mutants according the numbering of constructs described in A. Neurons were double immunostained for CLN6 (green) and the ER marker PDI or the Golgi marker GM130 (red). Yellow indicates overlap of CLN6 protein and subcellular markers. The square at the right is a magnified view of the indicated region. Insets represent 8-fold higher magnification images of the regions marked by the white rectangles. Bars, 5 µm. This Figure is reproduced in colour in Molecular Membrane Biology online.
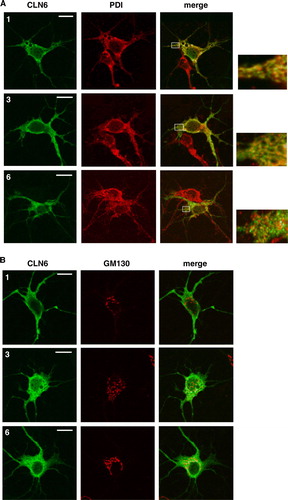
Figure 7. CLN6 forms homodimers. (A) BHK21 cells were transfected either with the empty vector, or with cDNAs coding for full-length CLN6, CLN6myc, or both CLN6 and CLN6myc. After 24 h the cells were permeabilized and treated (+) or not (−) with 0.5 mM BS3. The cell extracts were separated by SDS-PAGE and analysed by Western blotting using anti-CLN6-specific 1747 antibody. The positions of the molecular mass marker proteins and the different dimeric CLN6 forms are indicated. (B) BHK21 cells overexpressing either wild type CLN8, CLN6myc or both CLN8 and CLN6myc were permeabilized and treated with (+) or without (−) BS3. The cell extracts were separated by SDS-PAGE and analysed first by Western blotting using anti-CLN8 antibody. Thereafter, the blot was stripped and reprobed with anti-myc antibody. Vector-transfected cells were used as control. The blots were densitometrically scanned and the percentage of dimeric forms formed by cross-linkage are given.
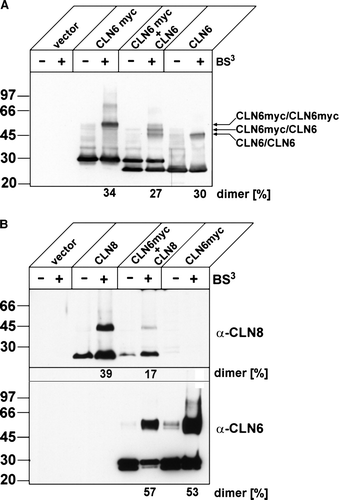
Figure 8. Capability of truncated CLN6 to dimerize. In BHK21 cells overexpressing wild type (1) or the indicated CLN6myc truncation mutants (p.Glu2_Leu20del = 2; p.Glu2_Phe49del = 3; p.Glu2_Ile118del = 4; p.Leu286X = 5; p.Glu225X = 6) membranes were permeabilized and proteins chemically cross-linked with (+) or without (−) 0.2 mM BS3. The cell extracts were separated by SDS-PAGE followed by Western blot analysis using anti-myc antibodies. The blots were densitometrically scanned and the percentage of dimeric forms formed by cross-linkage are given
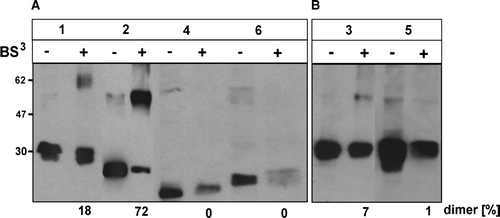
Table I. Relative expression of dimeric CLN6 forms.