Figures & data
Table 1. Primers used in this study.
Figure 1. Alignment of the amino acid sequences of olive flounder RH isolated in this study with those from the GenBank/EMBL/DDBJ databases. The GenBank accession numbers for the sequences used in the alignment are as follows: olive flounder (ofRH, HQ413772), Atlantic halibut (ahRH, AAM17918), common sole (csRH, CAA77254), flathead mullet (fmRH, CAA77250) and winter flounder (wfRH, AAT72123). The seven transmembrane domains are indicated by boxes. Amino acid residues involved in Schiff base formation (Phe, □) and their counterions (Thr, ▪), those involved in disulphide bond formation (Cys, *), those involved in glycosylation (glutamate residue; Gly, Δ) and those involved in palmitoylation (Asn and Pro, •) are indicated. Amino acids involved in the spectral tuning of rhodopsin are also indicated (Ala, ○).
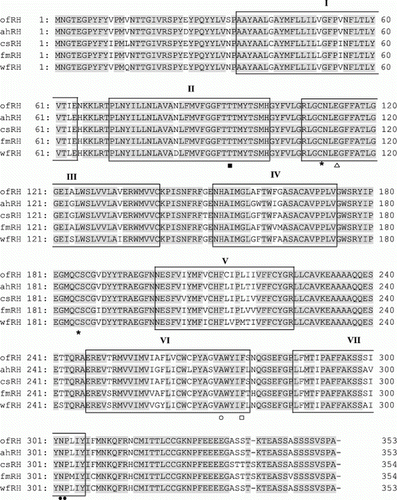
Figure 2. Alignment of the amino acid sequences from olive flounder Exo-RH isolated in this study with those from the GenBank/EMBL/DDBJ databases. The GenBank accession numbers for the sequences used in the alignment are as follows: olive flounder (ofExo-RH, ADI59669), fire clownfish (fcExo-RH, ADI59664), Fugu rubripes (frExo-RH, AF201472), ayu (ayExo-RH, BAC56700) and Atlantic salmon (asExo-RH, AAF44619). Putative transmembrane domains I–VII are indicated by horizontal lines. The conserved functional opsin features include glycosylation sites (Asn, Δ), a disulphide bridge (Cys, *), a chromophore attachment site (Lys, □), a Schiff base counterion (Glu, ▪); and a palmitoylation site (Asn and Pro, •).

Figure 3. Phylogenetic tree based on an amino acid alignment for RH and Exo-RH sequences in teleost fish. Bootstrap values (%) are indicated (1000 replicates). The score between two protein sequences, which is a measure of their relative phylogenetic relationship, is represented by the horizontal distance in this tree, i.e. the shorter the distance, the more related they are. GenBank accession numbers for the sequences are: Atlantic halibut RH (ahRH, AAM17918); common sole RH (csRH, CAA77254); flathead mullet RH (fmRH, CAA77250); winter flounder RH (wfRH, AAT72123); fire clownfish Exo-RH (fcExo-RH, ADI59664); Fugu rubripes Exo-RH (frExo-RH, AF201472); ayu Exo-RH (ayExo-RH, BAC56700); and Atlantic salmon Exo-RH (asExo-RH, AAF44619).
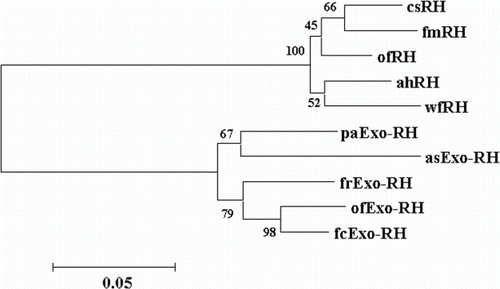
Figure 4. Tissue distribution of RH and Exo-RH mRNA in olive flounder. β-actin mRNA was amplified to verify the integrity of each mRNA sample.
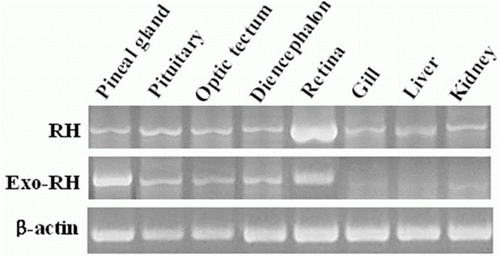
Figure 5. Diurnal variations in the levels of RH mRNA in the retina of the olive flounder as measured by quantitative real-time PCR. The fish were reared under a 12:12 light:dark (LD) cycle (a), constant dark (DD) (b) and constant light (LL) (c). Total retinal RNA (2.5 g) was reverse transcribed and amplified. The results are expressed as normalised expression levels with respect to the β-actin and GAPDH levels in the same sample. The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n = 5).
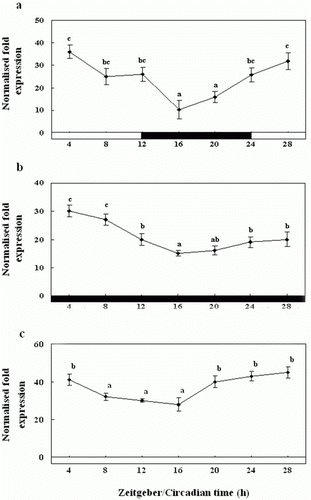
Figure 6. Diurnal variations in the levels of Exo-RH mRNA in the pineal gland in vivo as measured by quantitative real-time PCR. The fish were reared under a 12:12 light:dark (LD) cycle (a), constant dark (DD) (b) and constant light (LL) (c). Each mean value and error bar indicates the pineal gland from 10 fish. Total pineal gland RNA (2.5 g) was reverse transcribed and amplified. The results are expressed as the normalised expression levels with respect to the levels of β-actin and GAPDH in the same sample. The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n = 5).
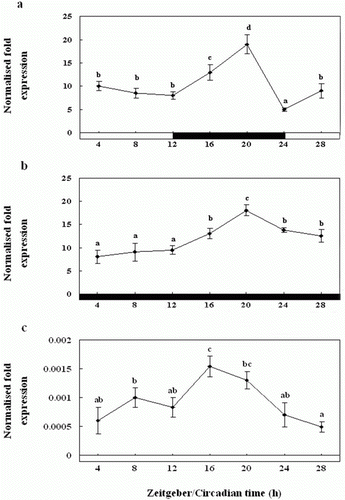
Figure 7. Diurnal variations in the levels of Exo-RH mRNA in the cultured pineal gland in vitro as measured by quantitative real-time PCR. The pineal gland was maintained under a 12:12 light:dark (LD) cycle (a), constant dark (DD) (b) and constant light (LL) (c). Each mean value and error bar indicates the pineal gland from 10 fish. Total pineal gland RNA (2.5 g) was reverse transcribed and amplified. The results are expressed as the normalised expression levels with respect to the levels of β-actin and GAPDH in the same sample. The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n = 5).
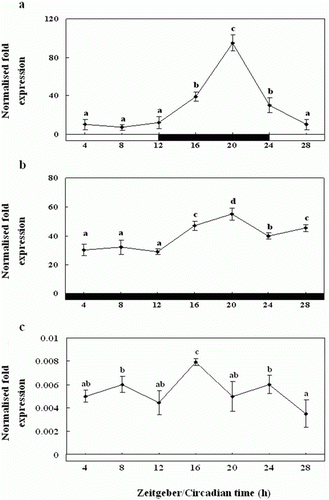
Figure 8. Diurnal variations in the levels of Exo-RH mRNA in the cultured melatonin-treated in vitro, as measured by quantitative real-time PCR. The pineal gland was maintained under a 12:12 light:dark (LD) cycle (a), constant dark (DD) (b) and constant light (LL) (c). Each mean value and error bar indicates the pineal gland from 10 fish. Total pineal gland RNA (2.5 g) was reverse transcribed and amplified. The results are expressed as the normalised expression levels with respect to the levels of β-actin and GAPDH in the same sample. The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n=5).
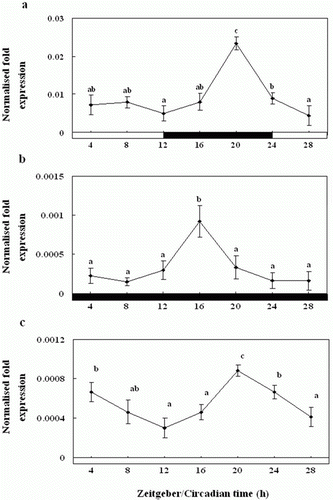
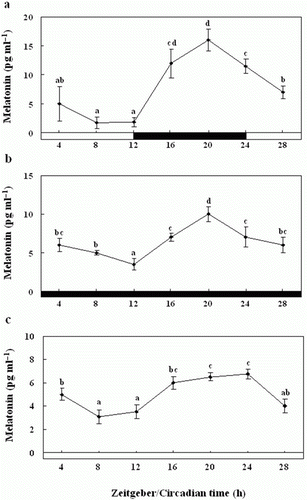
Figure 9. Enzyme-linked immunosorbent assay of the melatonin levels of the pineal gland culture medium during the daily light:dark (LD) cycle (12:12) (a), constant dark (DD) (b) and constant light (LL) (c). The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n = 5).
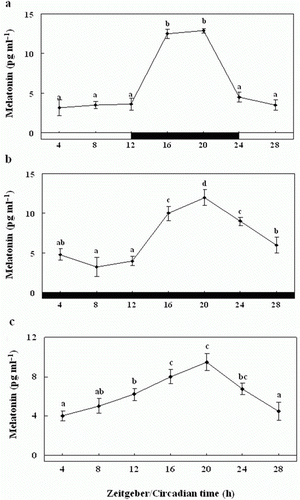
Figure 10. Enzyme-linked immunosorbent assay of plasma melatonin levels of olive flounders during the daily light:dark (LD) cycle (12:12) (a), constant dark (DD) (b) and constant light (LL) (c). The white bar represents the photophase and the black bar, the scotophase. Different letters indicate that values are statistically different in Zeitgeber time (ZT) and Circadian time (CT) (p < 0.05). All values represent means±SD (n = 5).