Figures & data
Figure 1. The hepatocytes incubating with (A) and without (B) QBT, observed in the microscope (×10).
Table 1. Primer sequence for molecular cloning and quantitative real-time PCR of FAT/CD36 in grass carp (C. idellus). All sequences are presented as 5′-3′.
Table 2. Composition and nutrient levels of the experimental diets (air-dry basis, %) for grass carp (C. idellus) used in the in vivo study.
Table 3. FA composition of diets (% of total FA) used in the in vivo study with grass carp (C. idellus).
Figure 2. Time-course uptake of LCFAs in hepatocytes determined by means of the QBT kit from grass carp (C. idellus).
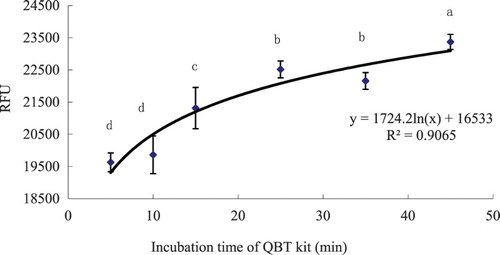
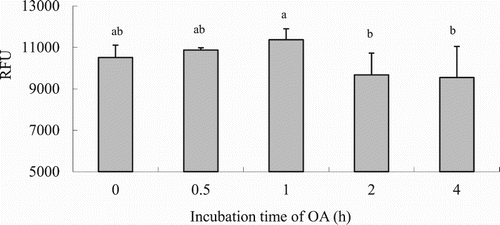
Figure 3. LCFA uptake depending on the OA concentration in hepatocytes from grass carp (C. idellus).
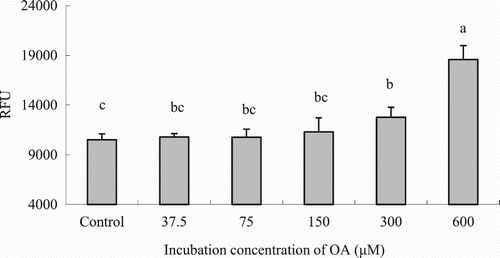
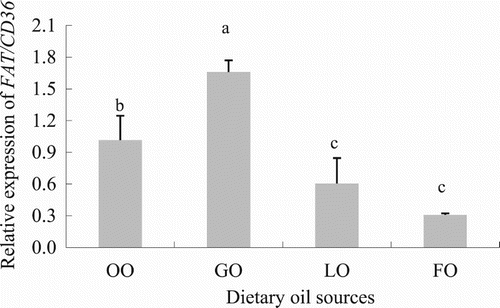
Figure 5. Effects of LCFA uptake depending on the type of FA in hepatocytes from grass carp (C. idellus).
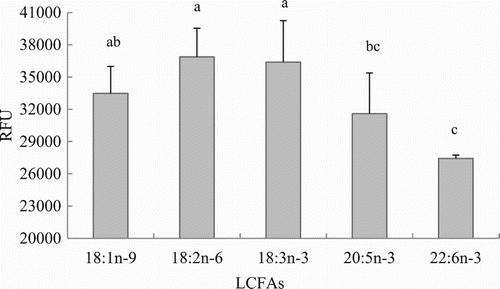
Figure 6. Effects of non-specific inhibitors on LCFA uptake in hepatocytes from grass carp (C. idellus).
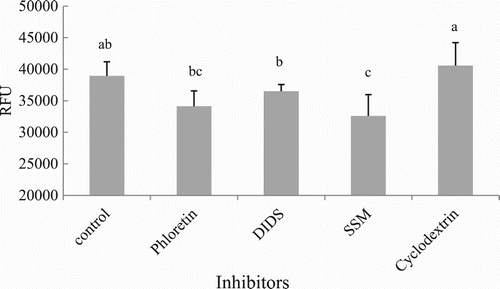
Figure 7. Comparison of FAT/CD36 amino acids’ homology in grass carp (C. idellus), common carp (C. carpio) and zebrafish (D. rerio). The line of consensus shows the conserved residues of FAT/CD36 among these fish and these identical amino acid residues are shown in white words with black background. The trans-membrane domain of FAT/CD36 is boxed. The alignment was generated using vector DNAMAN software.
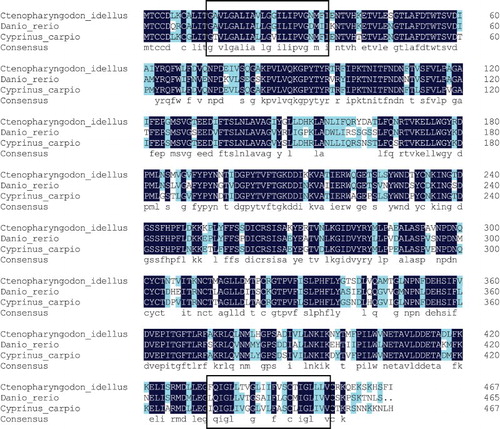
Figure 9. Secondary structure of FAT/CD36 protein is predicted. The domain of amino acids in blue line are α-helices, the domain of amino acids in red line are β-brands and the amino acids left in purple line are random coils.
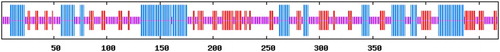
Figure 10. Three-dimensional model of FAT/CD36 shown by cartoon and surface models. This protein included many α-helices, β-brands and random coils. It was generafted by PHYRE2 Server.
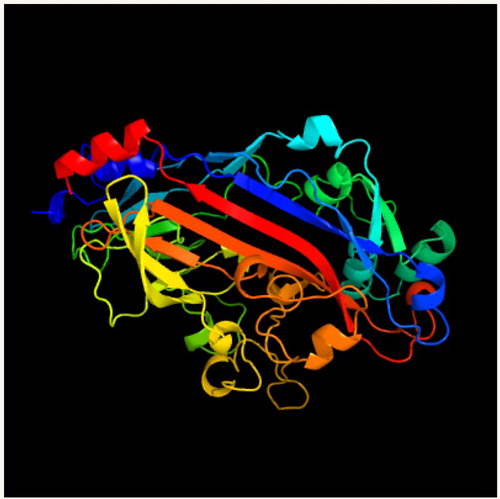
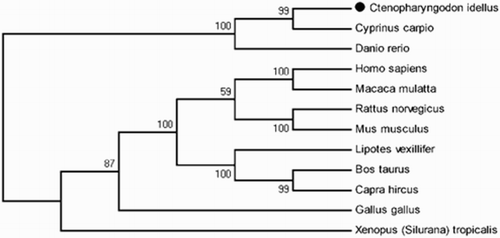
Figure 11. Phylogenetic analysis of FAT/CD36. Phylogenetic tree based on protein sequences was constructed by the neighbour-joining method with Mega 5.0 software. The strength of branch relationships was assessed by bootstrap replication (N = 1000 replicates). Grass carp FAT/CD36 was indicated by ‘●’. Accession numbers of protein sequences for FAT/CD36 are as follows: Cyprinus carpio (KM030422); Danio rerio (NM_001002363); Homo sapiens (KR710356); Macaca mulatta (AY600441); Rattus norvegicus (NM_031561); Mus musculus (NM_001159558); Lipotes vexillifer (XM_007467039); Bos taurus (NM_001278621); Capra hircus (JF690773); Gallus gallus (NM_001030731); Xenopus (Silurana) tropicalis (NM_001113679).