Figures & data
Figure 1. DNA sequence alignment of the UL36 partial open reading frame. The indel region is indicated by a 27-nt insertion (at a copy number of 2–5 presented as boxes outlined with black or red, and the background of blue, yellow, purple, and grey). The differentiation of the BHV-1.2 allele vs. BHV-1.1 allele is indicated by some degeneracy due to point mutation of C to T in the indel (red block letter, in sample B/1; B/31; B/32, and L/33). The dashes indicate spaces introduced to provide alignment among fragments of different lengths (due to the presence/absence of the indel).

Figure 2. (a) Sequence alignment of deduced amino acid (aa) sequence of the gD protein, extending from aa 137– 306 of the full-length protein. Arrows indicate the positions of aa residues corresponding to the SNPs. Blue and red backgrounds indicate amino acids that differ between BHV-1.1 and BHV-1.2 (respectively). (b) Sequence alignment of the deduced aa sequence of the UL36 protein extending from aa 2428–2496 of the full-length protein. The arrow indicates the position of the aa residue that differs between BHV-1.1 and BHV-1.2. The red-outlined box indicates the sequence region corresponding to the indel. The black box indicates the sequences that are unique to BHV-1.2 as a result of the indel and the changed amino acid residue. Green, blue, yellow, and purple backgrounds indicate the positions of the repeated 9-aa peptide sequence encoded by the indel.
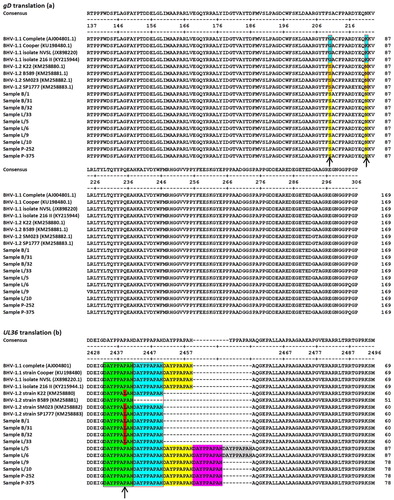
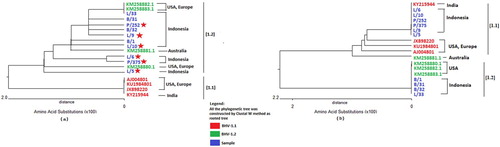
Figure 3. (a) Phylogenetic tree based on analysis of amino acid sequences encoded by the gD ORF fragment. Red indicates the BHV-1.1 group. Green indicates BHV-1.2, and blue indicates samples characterized in the present study. Red stars indicate the sample isolates that move from one group to another between the trees based on DNA vs. protein sequences. (b) Phylogenetic tree based on analysis of amino acid sequences encoded by the UL36 ORF fragment.