Figures & data
Table 1. Primers used for nucleotide sequencing of full-length Tembusu virus polyprotein gene.
Figure 1. Egg production rate (line) and the number of daily deaths (vertical bars) of the breeder Pekin ducks in the farm infected with Tembusu virus. A decrease in egg production was first observed on August 22, 2019. Carcasses of diseased ducks were submitted for disease investigation on September 4. Without intensive intervention, the drop in egg production and mortality recovered gradually after mid-September.
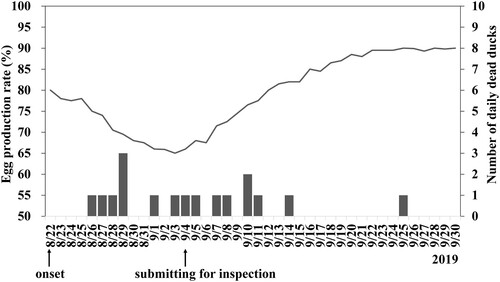
Figure 2. Negative staining electron micrograph of allantoic fluid harvested from 9-day-old embryonated Muscovy duck egg infected with Tembusu virus 1080905. Spherical virions with diameters of approximately 45–50 nm were observed.
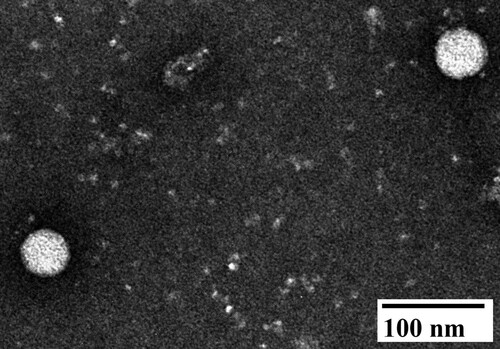
Figure 3. Phylogenetic analysis of the Tebmusu virus (TMUV) isolate TMUV 1080905 and 47 selected reference TMUV strains and duck TMUV based on the polyprotein gene (10,287 bp). The phylogenetic tree was constructed using the maximum likelihood method based on the Tamura-Nei model (Tamura and Nei Citation1993). Only bootstrap values (after 1000 replicates) over 70% are indicated at each branch point as a percentage. For each virus strain, strain name, host, year of isolation or detection and GenBank accession number are shown.
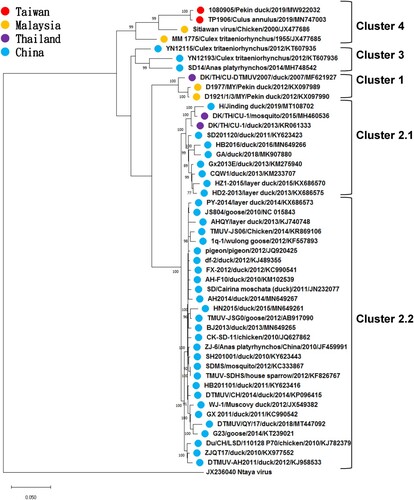
Table 2. The similarity of polyprotein gene and its encoded protein of Taiwanese Tembusu virus isolate TMUV 1080905 against other Tembusu viruses.
Table 3. Sequence comparisons of polyprotein of Tembusu virus isolates 1080905 and TP1906.
