Figures & data
Figure 1. The composition of DCN which included 245 nodes and 845 interactions. Yellow nodes stood for the ego genes.
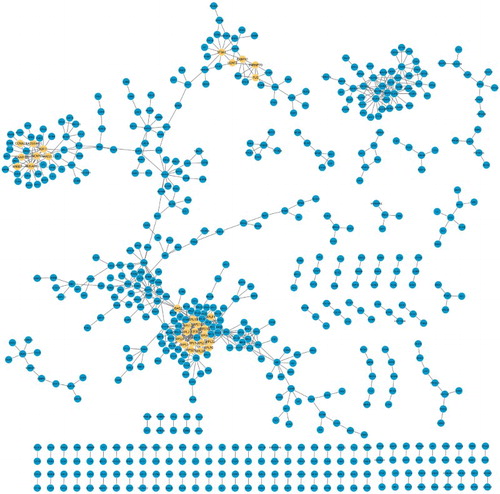
Table 1. The 30 ego genes extracted from the DCN and their z-scores.
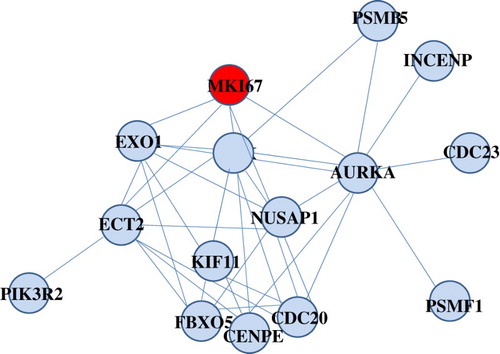
Figure 2. Composition of differential ego-network module 30 identified in the childhood ALL resistant to prednisolone treatment. Yellow node was ego gene.