Figures & data
Table 1. Hematological and biochemical parameters of four NADH-Cytb5R deficient Indian families.
Figure 1. Pedigree of the four families with RCM Type I disease due to a deficiency of the NADH-cytb5r enzyme associated with novel p.Arg192 > Cys mutation.

Figure 2. (A) Electropherogram of the exon 7 sequence indicating the T to C transition in position 22490 of the wild-type CYB5R3 sequence (OMIN:250800) and, top, the Box amino acid exchange in codon 192. (B) Ethidium bromide-stained 2% agarose gel of PCR fragments from all family members. The T→C transition eliminates a single BstUI site in the PCR fragment of exon 7 (612 bp), Normal (255, 185, 172 bp), Mutant (427 and 185 bp) and heterozygous (427, 255, 185, 172 bp), which permits rapid screening for the new R192C mutation in CYB5R3 gene by a simple BstUI restriction digest without the need of DNA sequencing.
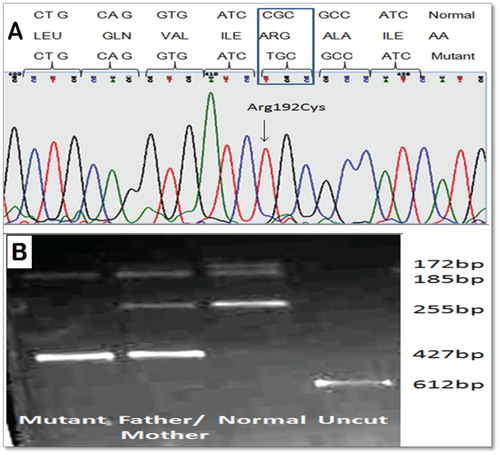
Figure 3. (A) Three-dimensional view of human NADH-cytochrome b5 reductase according to the crystallographic structure (7–8) In relation to the active site and the activation domain, the arginine in codon 192 (yellow) is found at the opposing surface or front side of the molecule and in close proximity to the NADH-binding domain. A cofactor FAD is non-covalently bound in a large and wide boundary cleft between the two domains. (B) and (C) show that Normal and the mutation of Arg-192 to Cys would be predicted to change the hydrogen bonds’ pattern between the regions of codon Ile 97 in the FAD-binding domain.

