Figures & data
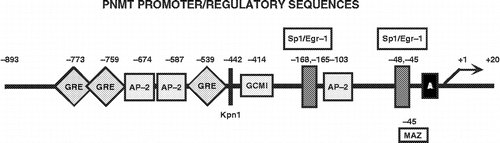
Figure 1 Catecholamine Biosynthesis. Depiction of steps involved in catecholamine biosynthesis. The amino acid tyrosine is converted to dihydroxyphenylacetic acid by TH. Dihydroxyphenylacetic acid is decarboxylated by dihydroxyphenylacetic acid decarboxylase (DDC) to the neurotransmitter dopamine. Addition of a hydroxyl group to the β carbon of the aliphatic chain by dopamine β‐hydroxylase (DBH) generates norepinephrine, and finally, norepinephrine is converted to epinephrine by N-methylation of the terminal amino group by PNMT.
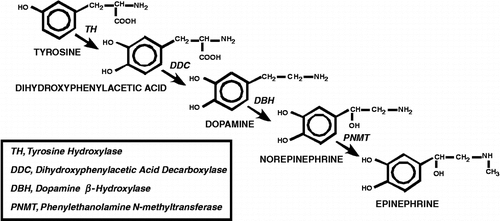
Figure 2 Tyrosine hydroxylase (TH) promoter. Schematic of the rat TH promoter depicting known transcription factor regulatory elements identified to date. The regulatory elements that are likely involved in the stress response are shown above the line: AP1 (activator protein-1 site that recognizes Jun and Fos proteins; Kumer and Vrana Citation1996); Sp1/Egr1 site (Papanikolaou and Sabban Citation1999) and the CRE (cAMP/CRE that recognizes members of the CREB, CREM and ATF families; Lewis-Tuffin et al. Citation2004). The sites designated below the line are involved in tissue-specific or developmental expression of the gene or responses to more specific stimuli. These include sites for hypoxia-inducible factor (HIF), AP2, POU domain factors, cAMP and TPA response factors (CRE2) and Nurr1.
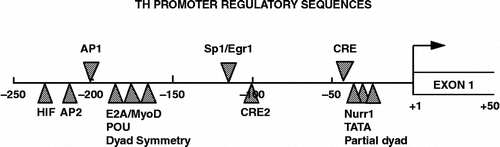
Figure 3 Phenylethanolamine N-methyltransferase (PNMT) Promoter. Schematic of the rat PNMT promoter depicting known transcription factor regulatory elements identified to date. Transcriptional activators include Egr-1 (Ebert et al. Citation1994; Ebert et al. Citation1998) and Sp1 (Ebert and Wong Citation1995) with two overlapping consensus sites at − 165/ − 168 and − 45/ − 48 bp upstream of the site of transcription initiation (+1), the GR (Ross et al. Citation1990; Tai et al. Citation2002) with consensus sites at − 539 and two overlapping sites at − 759/ − 773 bp, AP-2 (Ebert et al. Citation1998) with three consensus sites at − 103, − 587 and − 674 bp, MAZ (Her et al. Citation1999; Her et al. Citation2003) with consensus site at − 45 bp and GCM (Tai and Wong Citation2003) with consensus site at − 414 bp.