Figures & data
Figure 1. A model depicting the positive feedback loop between multi-omics data generation and isolation of as yet uncultured microorganisms. The rise of multi-omics tools has led to a better understanding of microbial life in nature, the resource for novel biotechnological applications needed by our society. The tedious cultivation of microorganisms often represents the first milestone in novel biotechnological process development and facilitates testing of ecological hypotheses. Multi-omics information, curated by physiological characterization of already available microbial isolates, represents a huge pool of knowledge about the yet-uncultured microbial world. Hence, integrating multi-omics data directed at culturing novel environmental bacteria (dashed arrow) brings multi-omics data into context and has the potential to boost biotechnological innovation for the benefit of society and nature conservation.
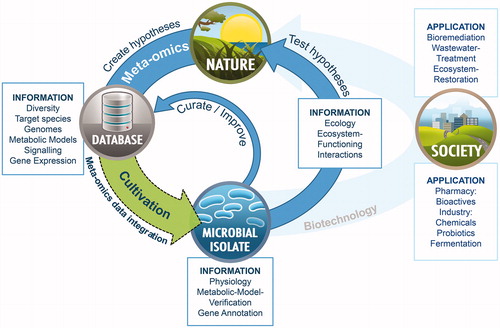
Figure 2. Schematic depiction of selected environments discussed in this review according to the diversity of the residing microbial community.
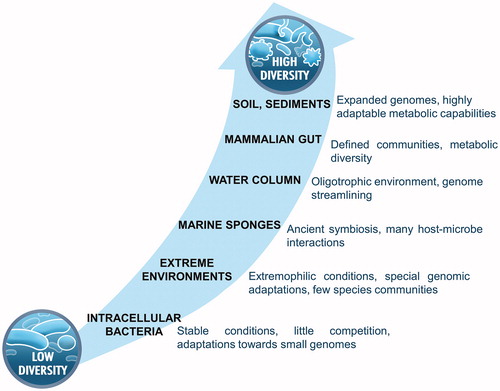
Figure 3. Proposed workflow for integrating multi-omics data into microbial cultivation. Arrows indicate the flow of information, blue for multi-omics strategies and green for microbial cultivation (in correspondence to ). *The sampling strategy prior to metagenomic or single-cell genomics differs for low- and high-diversity environments, the latter requiring a larger number of samples for enhanced genome recovery.
Table 1. Summary of existing literature on the use of multi-omics for isolating as-yet uncultured microorganisms.
