Figures & data
Figure 1. Proviral organization of HML-2 and transcripts. In the proviral form of HML-2, the sequences of the four ORFs overlap (shown in the scheme by the colored lines). Splice donor (SD) and splice acceptor (SA) sites are shown. LTRs are composed of the U3 and U5 regions separated by the R segment. As opposed to canonical retroviruses, which include the R segment in their transcripts, HML-2 transcription starts after the R. Transcript 1 has three ORFs to encode proteins GAG, PRO and POL. In this transcript, only gag has a start codon (AUG); pro and pol translation is mediated by two ribosomal frame shifts (–1). As a result, and despite having overlapping DNA sequences, the three proteins do not have any amino acid sequence in common. The figure also shows the ORFs organization to encode the different final proteins and domains (matrix (MA), capsid (CA) and nucleocapsid (NC) in gag, dUTPase in Gag-Pro junction and reverse transcriptase (RT), Rnase H and integrase (IN) in pol). The functional proteins will be formed by proteolysis of the polyproteins Gag, Gag-Pro and Gag-Pro-Pol. Transcript 2 encodes the protein Env, which has three different domains: the signal peptide (SP), surface (SU) and transmembrane (TM). Transcript 3 is the product of alternative splicing of the env ORF and encodes Rec. This transcript is only produced by type 2 HML-2 proviruses, which do not have the 292 nts deletion. Rec has 87 amino acids in common with Env, corresponding to its first exon. The second exon starts in a different frame and therefore, the amino acid sequence is not shared with Env. Transcript 4 is only produced by type 1 proviruses, which contain a deletion of 292 nts in the pol–env junction. As a result, the SD2 site is lost and an alternative SD (SDNP9) is used for the splicing. Due to this change only the first 14 amino-acids of NP9 are shared with Env and Rec.
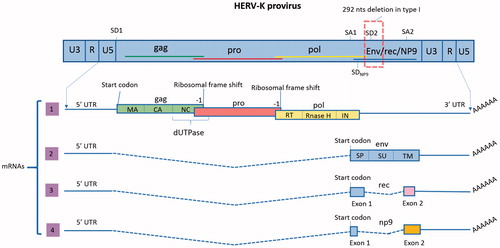
Table 1. HML-2 proviruses with ORFs for the main viral genes and some domains.
Table 2. Transcription factors binding to different HML-2 LTRs.
Figure 2. Molecular biosynthesis of HML-2. In the human genome HML-2 sequences can be found in the form of two types of proviruses (type 1 and type 2) as well as solo LTRs, resulting from homologous recombination between the two LTRs of a provirus and subsequent loss of the internal sequence. The two types of proviruses differ in the presence (type 1) or absence (type 2) of a 292 nts-deletion in the pol–env junction. Because of such deletion, type 1 proviruses are unable to encode the Env protein and the accessory protein Rec, encoding NP9 instead. In addition to transcripts with coding capability, non-coding RNAs are produced, resulting from the transcription of loci with premature stop codons. The consensus sequence of HML-2 provirus (type 2) presents a 5′LTR, a PBS and four main ORFs: (1) gag, encoding the structural proteins: matrix (MA), capsid (CA) and nucleocapsid (NC); (2) protease (pro), also encoding the enzyme dUTPase; (3) polymerase (pol), encoding the reverse transcriptase (RT), RNase H and integrase (IN) and (4) Envelope (env); a polypurine tract (PPT) and a 3′LTR. HML-2 gag, pro and pol transcripts are translated as polyproteins. The position in the reading frames indicates that ribosomal frameshifting is performed to synthesize Gag–Pro and Gag–Pro–Pol polyproteins. After autocleavage, the viral protease (Pro) will proteolyze the precursors, yielding the mature structural proteins of the matrix (MA), capsid (CA) and nucleocapsid (NC), and the active enzymes reverse transcriptase (RT) and integrase (IN). Env transcript can undergo alternative splicing, generating either env or rec/np9 mRNAs (depending on the type (1 or 2) of provirus). HML-2 Env is synthesized as a polyprotein that follows the secretory pathway. It has a signal peptide (SP) to direct the protein to the endoplasmic reticulum, where it is cleaved by the signal peptidase. Then Env is cleaved by furin host proteases into a surface unit (SU), and a transmembrane unit (TM). SU and TM are non-covalently associated and will possibly trimerize in the Golgi apparatus, resulting in a trimer of heterodimers. Env anchors into the cell membrane via the TM subunit, then traffics to the plasma membrane and studs the surface of the newly budding virus particles.
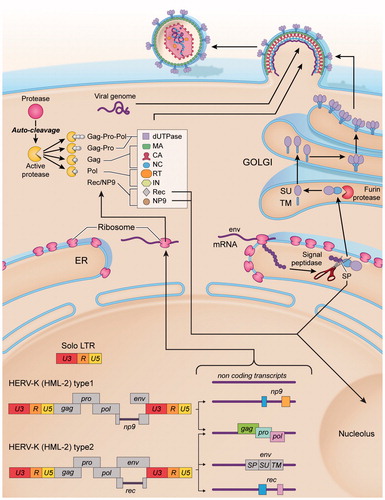
Figure 3. Potential mechanisms for HML-2-induced carcinogenesis. (A) Chromosomal rearrangements: HML-2 LTRs may contribute to carcinogenesis by facilitating homologous recombination, resulting in deletions, duplications, inversions or fusions of interspersed genomic sequence. (B) LTR-induced upregulation of adjacent oncogenes: If the LTR of HML-2 is de-repressed, it could recruit transcription factors and serve as an alternative promoter of adjacent host genes involved in cell proliferation. (C) HML-2-derived oncoproteins: NP9 and Rec contribute to cellular proliferation, by activation of ERK, AKT and Notch1, c-myc and beta-catenin. HML-2 Env has been shown to promote cell transformation and increase cellular migration and invasion, which could lead to metastasis. (D) HML-2 immunosuppression: NP9 and Rec can upregulate beta-catenin, which induce immune tolerance to tumours. HML-2 Env has an immunosuppressive domain in the TM subunit that triggers upregulation of IL-10.
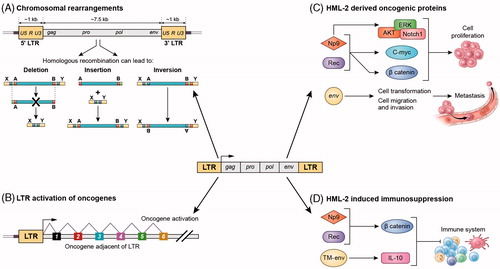
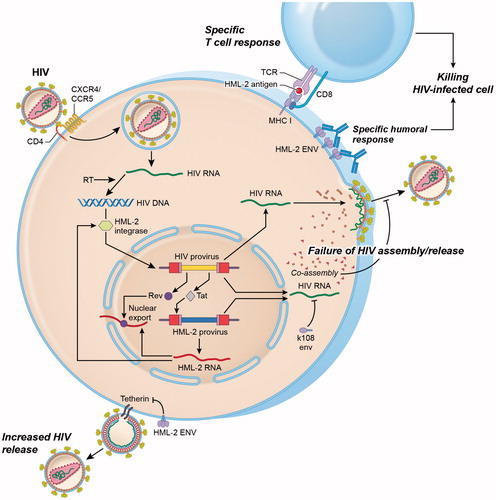
Figure 4. Interactions between HML-2 and HIV. Productive infection with HIV upregulates HML-2 expression at the mRNA level. Upregulation occurs at least partly as a result of the HIV regulatory protein Tat, which activates the NFkB and NF-AT pathways that enhance both HIV and HML-2 transcription. Similarly, HIV Rev binds to the Rev response element (RRE) in the env region of unspliced and partially spliced HIV transcripts to mediate nuclear export. HML-2 Env proteins may also be produced and expressed on the surface of HIV-infected lymphocytes. Increased levels of HML-2 Env-specific antibodies have been observed in HIV-positive individuals. HML-2-Gag and Env proteins may also be processed intracellularly for presentation in the context of MHC class I molecules, leading to activation of HML-2 specific cytotoxic T cell responses. Similarly, the HML-2 integrase can mediate integration of an integrase-deficient HIV provirus under in vitro conditions. HML-2 co-assembles with HIV Gag in vitro impairing HIV assembly and release. Expression of HML-2-Env from specific type 2 proviruses (K108, K109) also inhibits HIV release. In contrast, Env derived from a consensus HML-2 sequence can substitute for the HIV Vpu protein by downregulating the HIV restriction factor tetherin, leading to enhanced viral release.