Figures & data
Figure 1. Epigenetics and the mammalian life cycle. Following fertilization epigenetic marks are removed to generate a totipotent zygote, capable of producing all the different cell types required to form a new individual. Re-establishment of new epigenetic marks then redirects and drives development of the zygote through embryogenesis and ultimately into a mature adult. Primordial germ cells (PGCs) undergo a second epigenetic re-programing and programing to ensure correct imprinting and development of mature germ cells according to the gender of the embryo, and enable the successful production of subsequent generations in a continuous cycle. Such epigenetic processes can be perturbed by environmental factors, potentially leading to an adverse phenotypic outcome.
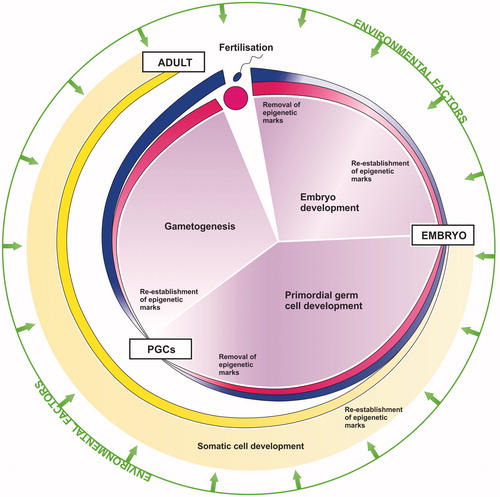
Figure 2. Epigenetic regulation of gene expression. A range of epigenetic processes including DNA methylation, histone modifications and non-coding RNAs (ncRNAs), (such as long ncRNAs (lncRNAs), microRNAs (miRNAs), piwi-interacting RNAs (piRNAs) and endogenous short interfering RNAs (esiRNAs)) form an efficient and robust complex interactive network that regulates gene expression at the transcriptional and post-transcriptional levels. Inhibitory processes are shown in red and positive in green. Both histone modifications and DNA methylation changes can inhibit or promote transcription of mRNAs and ncRNAs. ncRNAs can then inhibit the production of protein from mRNAs either via mRNA degradation and/or reduced translation, and/or protect genome stability via the degradation of transposable elements (retrotransposons). Some ncRNAs can also promote changes in DNA methylation, histone modifications, and/or the expression of other ncRNAs. To complete the network, DNA/RNA binding and modifying proteins can both inhibit or promote gene expression whilst their own expression is under the control of the DNA modifications and ncRNAs they promote and/or interact with.
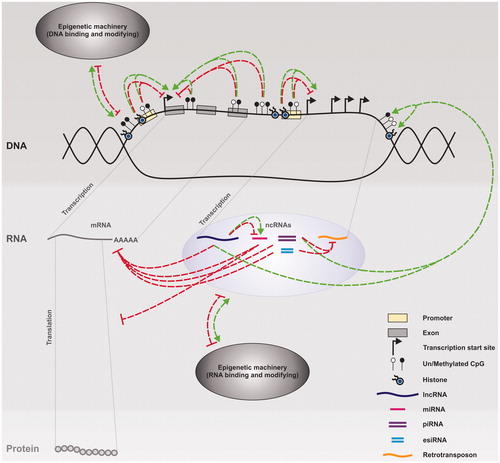
Figure 3. Multigenerational versus transgenerational epigenetic toxicity following in utero or ex utero environmental exposure. Penetration of adverse effect(s) to the first non-exposed generation (the F3 generation following in utero exposure, or the F2 generation following ex utero exposure) represents true epigenetic inheritance and thus transgenerational toxicity. Adverse effect(s) that affect multiple generations, all of which were exposed either directly or indirectly to the original factor represent epigenetic effect(s) and thus multigenerational toxicity.
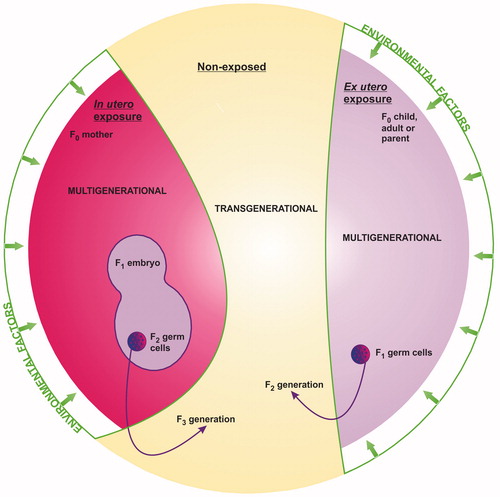
Table 1. Current evidence for putative environmentally induced epigenetic toxicity in human epidemiological cohorts.
Table 2. Current evidence for putative environmentally induced epigenetic toxicity in rodent models.
Table 3. Current OECD human health related TGs that have the potential for adaptation to include epigenetic endpoints.
