Figures & data
Figure 1. Structure of the Sleeping Beauty transposon system. (A) The Sleeping Beauty (SB) system. The transposase gene (yellow rectangle) is flanked by left and right inverted repeats (IRs) (arrows). Each IR contains two direct repeats (DR), an inner (DRI; orange) and an outer (DRO; brown) to which the transposase (yellow pie) binds at the respective core regions (gray thick line). (B) Domain organization of the transposase: The transposase consists of an N-terminal, DNA-binding domain (PAI + RED), a nuclear localization signal (NLS), an interdomain linker and a C-terminal, catalytic domain (CD). The CD has a clamp loop with a glycine strip (GGG) and three conserved catalytic residues (DDE). Both PAI and RED domains contain three alpha-helices and are separated from each other by a GRRR AT-hook motif. Numbers in the lowest panel represent residues that directly interact with the DNA. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
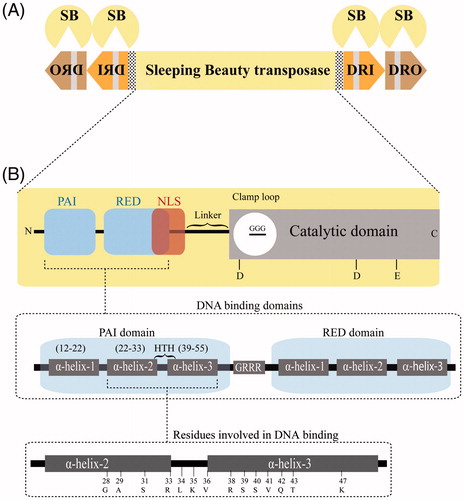
Figure 2. Host factors assist Sleeping Beauty transposition. Schematic representation of transposon mobilization: following expression (1) SB (red spheres) binds to the IRs (2) leading to the formation of a synaptic complex (3i) resulting in excision (3ii) from the donor DNA. The excised element then inserts into a TA dinucleotide (*) (3iii), which is duplicated following a successful integration. Excision would leave a 3-bp footprint behind. Role of host factors in regulating transposition: Transcriptional control: the high mobility group protein, HMGXB4 upregulates SB transcription (green arrow). Upon expression, the SB transposase antagonizes the effect of HMGXB4. The synaptic complex: SB recruits the high mobility group protein, HMGB1 (green ovals), which promotes synaptic complex assembly starting at the inner DRs. Prevention of autointegration: The host factor, barrier-to-autointegration factor (BAF1) prevents suicidal self-integration. Excision site repair: The Ku70/80 complex of the non-homologous end joining repair pathway (NHEJ) assists healing double stranded DNA damage generated upon transposon excision. Cell cycle modulation: SB modulates cell cycle transition via Miz1 by down regulating Cyclin D1 expression. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
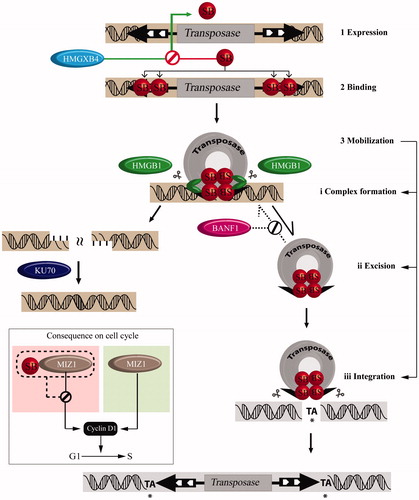
Figure 3. Structure of the Sleeping Beauty transposase. (A) Structure of the PAI domain as deciphered by NMR. The structure consists of three α helices (in orange; PDB 2M8E). Amino acids involved in DNA binding are highlighted in gray. (B) Superimposition of PAX5 (in green; PDB 1K78) with the PAI domain. The differences are highlighted by a dotted circle. (C) Structure of the catalytic domain (CD; in orange; PDB 5CR4) showing the N-terminal interdomain linker in gray, α helices in orange, β sheets in purple. The glycine strip (shown in cyan) is part of the clamp loop. The three catalytic residues, DDE are shown in gray. (D) Superimposition of the catalytic domain (CD) of SB (red) on MOS1 structure (cyan; PDB 3HOS). Note that main differences among them are located in the clamp loop (circled dotted line). (E) The potential role of the clamp loops in SB dimerization. (F) The mutation (I212S) further improves the hyperactivity of SB100X (Voigt et al., Citation2016). The DAVQ stretch is used as a reference to show the position of I-212 (refer the adjoining inlet). The structural analysis was done by the Chimera version of 1.10.2 (Pettersen et al., Citation2004). A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
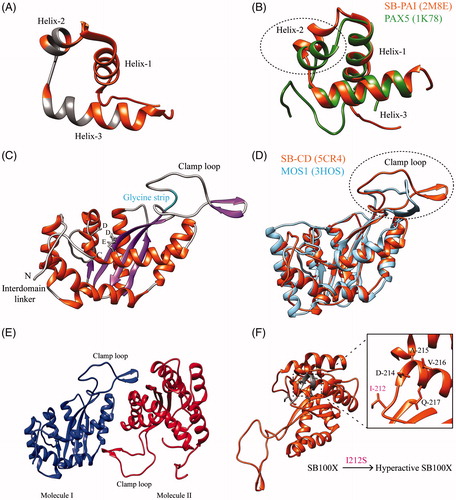
Figure 4. Integration profile of various integrating vectors. SB (red vertical lines) has a fairly random integration profile when compared to other DNA transposons like piggyBac (PB) (green vertical lines) and Tol2 (black vertical lines) and viral systems like HIV (cyan vertical lines) and MLV (green vertical lines). TSS: transcription start site. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
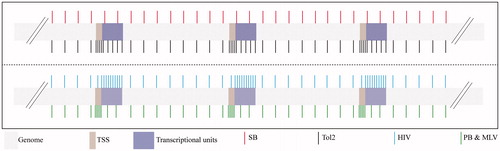
Figure 5. Sleeping Beauty transposon-based applications. SB was successfully used for germline transgenesis, in various models (fish1, frog2, rat3, mouse4, rabbit5, pig6, cow7 and sea squirt8). The SB system has been employed for somatic gene delivery in various vertebrates, but also in a tick (insect) cell line, ISE6. SB-based gene delivery has been used in several preclinical animal models. Alternatively, the mutagenic version of the SB can be employed in functional genomics. Insertional mutagenesis screens can be used to annotate genes in somatic cells (oncogenomics) or in the germline. Abbreviations: eNOS: endothelial nitric oxide synthase; hUGT1A1: human uridinediphosphoglucuronate glucuronosyltransferase-1A1; statin-AE: angiostatin–endostatin fusion cassette; DMD: Duchenne muscular dystrophy; DYSF: dysferlin; IDUA: α-L-iduronidase; FAH: fumarylacetoacetate hydrolase; INS: insulin; L/VDLRs: low-density lipoprotein and very-low-density lipoprotein receptors; miR–29: micro RNA 29; IOD: indoleamine-2,3 dioxygenase; DsRed2: red fluorescent protein 2; GFPs: green fluorescent proteins; siMSTN: siRNA against myostatin; siHTT: siRNA against Huntington; LAMB3: laminin subunit beta-3; HSVTK: herpes simplex virus thymidine kinase type 1 gene; BCP-ALL: B cell precursor acute lymphoblastic leukemia). A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
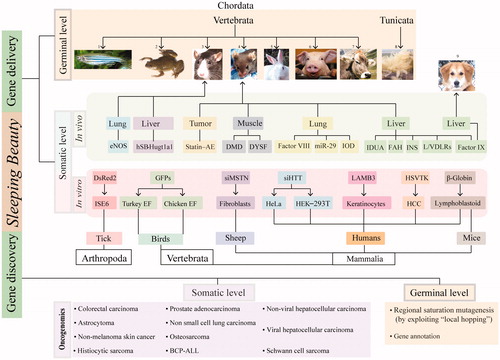
Figure 6. Multiple sequence alignment of Sleeping Beauty transposase sequences. Hyperactive amino acid mutations (in white and/or gray) compared to SB10 the original version of the SB transposase, SB10. Hyperactive SB11 and SB100X are targeted for clinical applications. Multiple sequence alignment was performed using EBI Clustal omega (Sievers et al., Citation2011) and shading was performed using BOXSHADE server version 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
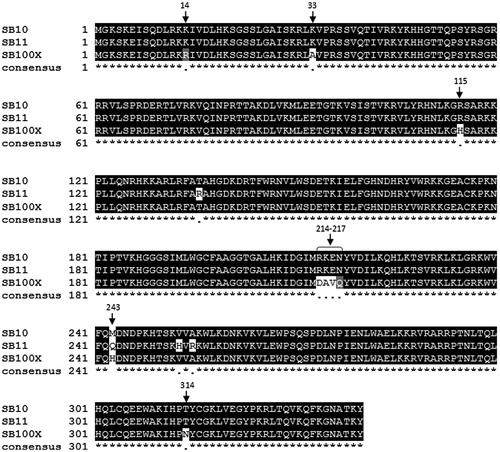
Figure 7. Modulating Sleeping Beauty target site specificity. Strategies to increase target site specificity of Sleeping Beauty integration. (A) Fusing a DNA-binding domain (DBD) (dark blue) to the N-terminus of the SB transposase (red). The affinity of DBD to DNA-binding domain region (DBDR) could direct transposon integration. (B) Co-delivery of a fusion construct with two DBDs, where DBD1 (cyan) binds an engineered region in the transposon (DBD1R, cyan), while DBD2 (orange) recognizes a genomic target sequence (DBD2R, orange). (C) Alternatively the SB system can be co-delivered with a a fusion protein, where a DBD (brown) is fused to an N-terminal, SB transposase derived peptide, N57 (green), a natural interaction partner of the full-length SB transposase. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
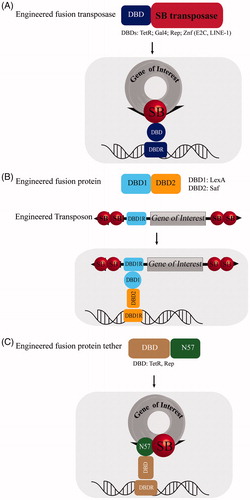
Figure 8. Sleeping Beauty transposon-based hybrid vectors. To circumvent the hindrance associated with transfection efficiency of naked DNA plasmids, the SB system, including both the transposase (Tnpase/SB – green dots for protein) and the transposon (Tnp/GOI – red) can be packaged into various recombinant viruses like Adeno, AAV, IDLV, HSV-1 and Baculovirus for delivery (by transduction) into the cytoplasm. The delivery is followed by quick and stable genomic integration of the cargo/GOI, mediated by the SB system as shown in follow-up qualitative time plot. Abbreviations: GOI: gene of interest; adeno: adenovirus; AAV: adeno associated virus; IDLV: integrase defective lentivirus; HSV-1: herpes simplex virus 1 amplicon; baculo: baculovirus. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).
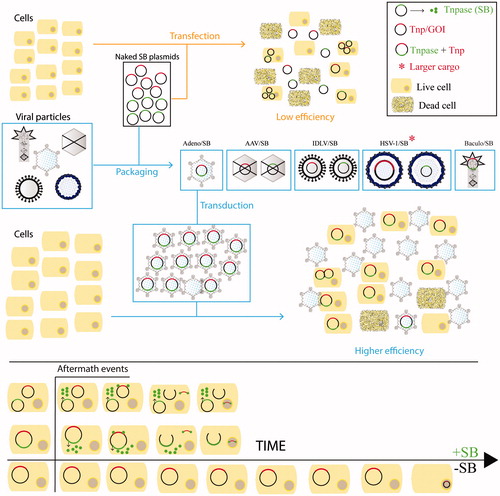
Table 1. Various sleeping beauty-viral hybrid technologies.
Table 2. Different types of viral vectors used for gene therapy.
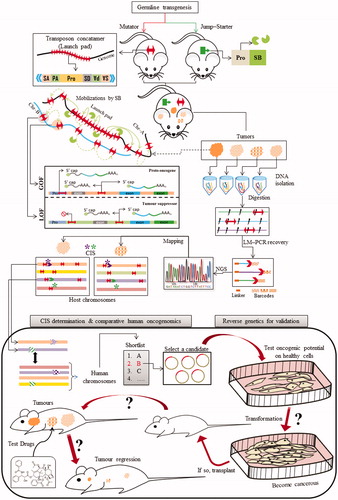
Figure 9. Sleeping Beauty transposon-based functional oncogenomics. An overall scheme depicting key steps for employing the SB system for oncogenomics. The conditional insertional mutagenesis begins with the generation of transgenic lineages of the chosen model system, popularly referred to as the mutator (transposon) and jump-starter (transposase) lineages. The mutator lineage is transgenic for a custom-engineered transposon (red double-headed arrow) that can be mobilized by the SB transposase (green pie). The mutator transposon is designed so that by virtue, it can promote, alter or even terminate expression of endogenous reading frames upon SB mediated insertion in either orientations eventually leading to gain and/or loss of functions. This entire scenario can be rendered “conditional” by restricting the activity of SB to specific tissues or organs of choice by placing it under the command of appropriate promoter. The double-transgenic animals are aged for development of relevant cancer phenotypes. The next step involves the identification of the transposon integration sites that very likely resulted in the observed phenotypes. This primarily involves extraction and digestion of the DNA from which these sites are recovered by LM-PCR, barcoded and sequenced by next generation sequencing. The recovered sites are mapped on the genome, and subjected to a statistical analysis to identify common integration sites (CISs) that were present in majority of the tumors. Shortlisted candidates are further validated by reverse genetic approach as illustrated in the figure. Abbreviations: SA: splice acceptor; PA: poly-A tail; pro: promoter; SD: splice donor; GOF: gain of function; LOF: loss of function; Chr: chromosome; LM-PCR: linker-mediated PCR; NGS: next generation sequencing; CIS: common integration sites. A color version of the figure is available online (see color version of this figure at www.informahealthcare.com/bmg).